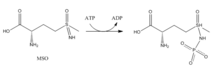
Glutamate dehydrogenase is an enzyme observed in both prokaryotes and eukaryotic mitochondria. The aforementioned reaction also yields ammonia, which in eukaryotes is canonically processed as a substrate in the urea cycle. Typically, the α-ketoglutarate to glutamate reaction does not occur in mammals, as glutamate dehydrogenase equilibrium favours the production of ammonia and α-ketoglutarate. Glutamate dehydrogenase also has a very low affinity for ammonia, and therefore toxic levels of ammonia would have to be present in the body for the reverse reaction to proceed. In the brain, the NAD+/NADH ratio in brain mitochondria encourages oxidative deamination. In bacteria, the ammonia is assimilated to amino acids via glutamate and aminotransferases. In plants, the enzyme can work in either direction depending on environment and stress. Transgenic plants expressing microbial GLDHs are improved in tolerance to herbicide, water deficit, and pathogen infections. They are more nutritionally valuable.
Biosynthesis, i.e., chemical synthesis occurring in biological contexts, is a term most often referring to multi-step, enzyme-catalyzed processes where chemical substances absorbed as nutrients serve as enzyme substrates, with conversion by the living organism either into simpler or more complex products. Examples of biosynthetic pathways include those for the production of amino acids, lipid membrane components, and nucleotides, but also for the production of all classes of biological macromolecules, and of acetyl-coenzyme A, adenosine triphosphate, nicotinamide adenine dinucleotide and other key intermediate and transactional molecules needed for metabolism. Thus, in biosynthesis, any of an array of compounds, from simple to complex, are converted into other compounds, and so it includes both the catabolism and anabolism of complex molecules. Biosynthetic processes are often represented via charts of metabolic pathways. A particular biosynthetic pathway may be located within a single cellular organelle, while others involve enzymes that are located across an array of cellular organelles and structures.

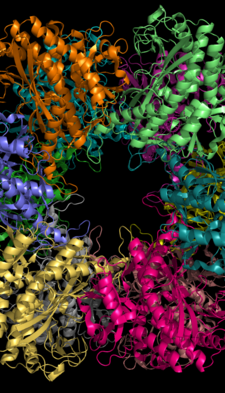
GLUD1 is a mitochondrial matrix enzyme, one of the family of glutamate dehydrogenases that are ubiquitous in life, with a key role in nitrogen and glutamate (Glu) metabolism and energy homeostasis. This dehydrogenase is expressed at high levels in liver, brain, pancreas and kidney, but not in muscle. In the pancreatic cells, GLUD1 is thought to be involved in insulin secretion mechanisms. In nervous tissue, where glutamate is present in concentrations higher than in the other tissues, GLUD1 appears to function in both the synthesis and the catabolism of glutamate and perhaps in ammonia detoxification.

Succinyl coenzyme A synthetase is an enzyme that catalyzes the reversible reaction of succinyl-CoA to succinate. The enzyme facilitates the coupling of this reaction to the formation of a nucleoside triphosphate molecule from an inorganic phosphate molecule and a nucleoside diphosphate molecule. It plays a key role as one of the catalysts involved in the citric acid cycle, a central pathway in cellular metabolism, and it is located within the mitochondrial matrix of a cell.
Carbamoyl phosphate synthetase I is a ligase enzyme located in the mitochondria involved in the production of urea. Carbamoyl phosphate synthetase I transfers an ammonia molecule to a molecule of bicarbonate that has been phosphorylated by a molecule of ATP. The resulting carbamate is then phosphorylated with another molecule of ATP. The resulting molecule of carbamoyl phosphate leaves the enzyme.

Amino acid biosynthesis is the set of biochemical processes by which the amino acids are produced. The substrates for these processes are various compounds in the organism's diet or growth media. Not all organisms are able to synthesize all amino acids. For example, humans can synthesize 11 of the 20 standard amino acids. These 11 are called the non-essential amino acids.

CAD protein is a trifunctional multi-domain enzyme involved in the first three steps of pyrimidine biosynthesis. De-novo synthesis starts with cytosolic carbamoylphosphate synthetase II which uses glutamine, carbon dioxide and ATP. This enzyme is inhibited by uridine triphosphate.
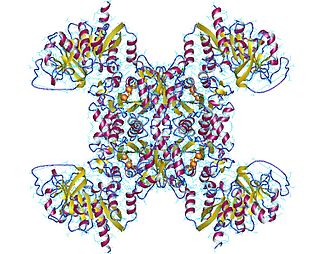
CTP synthase is an enzyme involved in pyrimidine biosynthesis that interconverts UTP and CTP.

Glutaminase is an amidohydrolase enzyme that generates glutamate from glutamine. Glutaminase has tissue-specific isoenzymes. Glutaminase has an important role in glial cells.

Carbamoyl phosphate synthetase catalyzes the ATP-dependent synthesis of carbamoyl phosphate from glutamine or ammonia and bicarbonate. This ATP-grasp enzyme catalyzes the reaction of ATP and bicarbonate to produce carboxy phosphate and ADP. Carboxy phosphate reacts with ammonia to give carbamic acid. In turn, carbamic acid reacts with a second ATP to give carbamoyl phosphate plus ADP.
Purine metabolism refers to the metabolic pathways to synthesize and break down purines that are present in many organisms.

Guanosine monophosphate synthetase, also known as GMPS is an enzyme that converts xanthosine monophosphate to guanosine monophosphate.

Amidophosphoribosyltransferase (ATase), also known as glutamine phosphoribosylpyrophosphate amidotransferase (GPAT), is an enzyme responsible for catalyzing the conversion of 5-phosphoribosyl-1-pyrophosphate (PRPP) into 5-phosphoribosyl-1-amine (PRA), using the amine group from a glutamine side-chain. This is the committing step in de novo purine synthesis. In humans it is encoded by the PPAT gene. ATase is a member of the purine/pyrimidine phosphoribosyltransferase family.
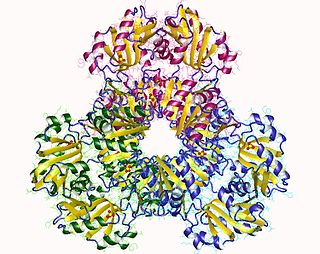
Ribose-phosphate diphosphokinase is an enzyme that converts ribose 5-phosphate into phosphoribosyl pyrophosphate (PRPP). It is classified under EC 2.7.6.1.

Adenylylation, more commonly known as AMPylation, is a process in which an adenosine monophosphate (AMP) molecule is covalently attached to the amino acid side chain of a protein. This covalent addition of AMP to a hydroxyl side chain of the protein is a post-translational modification. Adenylylation involves a phosphodiester bond between a hydroxyl group of the molecule undergoing adenylylation, and the phosphate group of the adenosine monophosphate nucleotide. Enzymes that are capable of catalyzing this process are called AMPylators.
Glutamine—tRNA ligase or glutaminyl-tRNA synthetase (GlnRS) is an aminoacyl-tRNA synthetase, also called tRNA-ligase. is an enzyme that attaches the amino acid glutamine onto its cognate tRNA.

The glutamine riboswitch is a conserved RNA structure that was predicted by bioinformatics. It is present in a variety of lineages of cyanobacteria, as well as some phages that infect cyanobacteria. It is also found in DNA extracted from uncultivated bacteria living in the ocean that are presumably species of cyanobacteria.

The PII family comprises a group of widely distributed signal transduction proteins found in nearly all Bacteria and also present in Archaea and in the chloroplasts of Algae and plants. PII form barrel-like homotrimers with a flexible loop, namely T-loop, emerging from each subunit. PII proteins have extraordinary sensory properties; they can exist in a vast range of structural status accordingly to the levels of ATP, ADP and 2-oxogluratate. These metabolites interact allosterically with PII in three conserved binding sites located in the lateral cavity between each PII subunit. ATP and ADP bind competitively to the nucleotide binding whereas the 2-oxoglutarate only interacts with PII in the presence of MgATP.
The glnALG operon is an operon that regulates the nitrogen content of a cell. It codes for the structural gene glnA the two regulatory genes glnL and glnG. glnA encodes glutamine synthetase, an enzyme which catalyzes the conversion of glutamate and ammonia to glutamine, thereby controlling the nitrogen level in the cell. glnG encodes NRI which regulates the expression of the glnALG operon at three promoters, which are glnAp1, glnAp2 located upstream of glnA) and glnLp. glnL encodes NRII which regulates the activity of NRI. No significant homology is found in Eukaryotes.
Carbamoyl phosphate synthetase III is one of the three isoforms of the carbamoyl phosphate synthetase, an enzyme that catalyzes the active production of carbamoyl phosphate in many organisms.