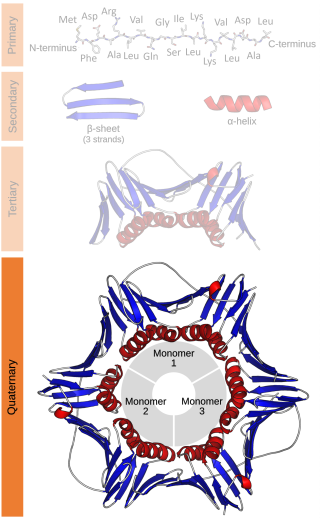
Protein quaternary structure is the fourth classification level of protein structure. Protein quaternary structure refers to the structure of proteins which are themselves composed of two or more smaller protein chains. Protein quaternary structure describes the number and arrangement of multiple folded protein subunits in a multi-subunit complex. It includes organizations from simple dimers to large homooligomers and complexes with defined or variable numbers of subunits. In contrast to the first three levels of protein structure, not all proteins will have a quaternary structure since some proteins function as single units. Protein quaternary structure can also refer to biomolecular complexes of proteins with nucleic acids and other cofactors.

A protein complex or multiprotein complex is a group of two or more associated polypeptide chains. Protein complexes are distinct from multidomain enzymes, in which multiple catalytic domains are found in a single polypeptide chain.
Propionic acidemia, also known as propionic aciduria or propionyl-CoA carboxylase deficiency, is a rare autosomal recessive metabolic disorder, classified as a branched-chain organic acidemia.

Acetyl-CoA carboxylase (ACC) is a biotin-dependent enzyme that catalyzes the irreversible carboxylation of acetyl-CoA to produce malonyl-CoA through its two catalytic activities, biotin carboxylase (BC) and carboxyltransferase (CT). ACC is a multi-subunit enzyme in most prokaryotes and in the chloroplasts of most plants and algae, whereas it is a large, multi-domain enzyme in the cytoplasm of most eukaryotes. The most important function of ACC is to provide the malonyl-CoA substrate for the biosynthesis of fatty acids. The activity of ACC can be controlled at the transcriptional level as well as by small molecule modulators and covalent modification. The human genome contains the genes for two different ACCs—ACACA and ACACB.
A dodecameric protein has a quaternary structure consisting of 12 protein subunits in a complex. Dodecameric complexes can have a number of subunit 'topologies', but typically only a few of the theoretically possible subunit arrangements are observed in protein structures.
A tetrameric protein is a protein with a quaternary structure of four subunits (tetrameric). Homotetramers have four identical subunits, and heterotetramers are complexes of different subunits. A tetramer can be assembled as dimer of dimers with two homodimer subunits, or two heterodimer subunits.

The enzyme argininosuccinate lyase (EC 4.3.2.1, ASL, argininosuccinase; systematic name 2-(N ω-L-arginino)succinate arginine-lyase (fumarate-forming)) catalyzes the reversible breakdown of argininosuccinate:
Holocarboxylase synthetase ), also known as protein—biotin ligase, is a family of enzymes. This enzyme is important for the effective use of biotin, a B vitamin found in foods such as liver, egg yolks, and milk. In many of the body's tissues, holocarboxylase synthetase activates other specific enzymes by attaching biotin to them. These carboxylases are involved in many critical cellular functions, including the production and breakdown of proteins, fats, and carbohydrates.

Methylmalonyl-CoA mutase is a mitochondrial homodimer apoenzyme that focuses on the catalysis of methylmalonyl CoA to succinyl CoA. The enzyme is bound to adenosylcobalamin, a hormonal derivative of vitamin B12 in order to function. Methylmalonyl-CoA mutase deficiency is caused by genetic defect in the MUT gene responsible for encoding the enzyme. Deficiency in this enzyme accounts for 60% of the cases of methylmalonic acidemia.

Biotin carboxyl carrier protein (BCCP) refers to proteins containing a biotin attachment domain that carry biotin and carboxybiotin throughout the ATP-dependent carboxylation by biotin-dependent carboxylases. The biotin carboxyl carrier protein is an Acetyl CoA subunit that allows for Acetyl CoA to be catalyzed and converted to malonyl-CoA. More specifically, BCCP catalyzes the carboxylation of the carrier protein to form an intermediate. Then the carboxyl group is transferred by the transcacrboxylase to form the malonyl-CoA. This conversion is an essential step in the biosynthesis of fatty acids. In the case of E. coli Acetyl-CoA carboxylase, the BCCP is a separate protein known as accB. On the other hand, in Haloferax mediterranei, propionyl-CoA carboxylase, the BCCP pccA is fused with biotin carboxylase.

Oxaloacetate decarboxylase is a carboxy-lyase involved in the conversion of oxaloacetate into pyruvate.

Methylmalonyl-CoA mutase (EC 5.4.99.2, MCM), mitochondrial, also known as methylmalonyl-CoA isomerase, is a protein that in humans is encoded by the MUT gene. This vitamin B12-dependent enzyme catalyzes the isomerization of methylmalonyl-CoA to succinyl-CoA in humans. Mutations in MUT gene may lead to various types of methylmalonic aciduria.
Propionyl-CoA is a coenzyme A derivative of propionic acid. It is composed of a 24 total carbon chain and its production and metabolic fate depend on which organism it is present in. Several different pathways can lead to its production, such as through the catabolism of specific amino acids or the oxidation of odd-chain fatty acids. It later can be broken down by propionyl-CoA carboxylase or through the methylcitrate cycle. In different organisms, however, propionyl-CoA can be sequestered into controlled regions, to alleviate its potential toxicity through accumulation. Genetic deficiencies regarding the production and breakdown of propionyl-CoA also have great clinical and human significance.

Methylmalonic acid (MMA) is a chemical compound from the group of dicarboxylic acids. It consists of the basic structure of malonic acid and also carries a methyl group. The salts of methylmalonic acid are called methylmalonates.
Methylcrotonyl CoA carboxylase is a biotin-requiring enzyme located in the mitochondria. MCC uses bicarbonate as a carboxyl group source to catalyze the carboxylation of a carbon adjacent to a carbonyl group performing the fourth step in processing leucine, an essential amino acid.

Methylmalonyl-CoA is the thioester consisting of coenzyme A linked to methylmalonic acid. It is an important intermediate in the biosynthesis of succinyl-CoA, which plays an essential role in the tricarboxylic acid cycle.
Carboxylation is a chemical reaction in which a carboxylic acid is produced by treating a substrate with carbon dioxide. The opposite reaction is decarboxylation. In chemistry, the term carbonation is sometimes used synonymously with carboxylation, especially when applied to the reaction of carbanionic reagents with CO2. More generally, carbonation usually describes the production of carbonates.

Methylmalonyl CoA epimerase is an enzyme involved in fatty acid catabolism that is encoded in human by the "MCEE" gene located on chromosome 2. It is routinely and incorrectly labeled as "methylmalonyl-CoA racemase". It is not a racemase because the CoA moiety has 5 other stereocenters.
In enzymology, a biotin carboxylase (EC 6.3.4.14) is an enzyme that catalyzes the chemical reaction
Yoshito Kaziro was a Japanese biochemical and medical scientist who performed research on the effects and mechanisms of ATP and GTP driven conformational changes in enzymes and intracellular signaling pathways for over 50 years. He is well-known for his research on various signal transduction pathways involving GTP-binding proteins and the mechanism for biotin dependent carboxylation reactions of Coenzyme A (CoA) proteins.
















