Leucine (symbol Leu or L) is an essential amino acid that is used in the biosynthesis of proteins. Leucine is an α-amino acid, meaning it contains an α-amino group (which is in the protonated −NH3+ form under biological conditions), an α-carboxylic acid group (which is in the deprotonated −COO− form under biological conditions), and a side chain isobutyl group, making it a non-polar aliphatic amino acid. It is essential in humans, meaning the body cannot synthesize it: it must be obtained from the diet. Human dietary sources are foods that contain protein, such as meats, dairy products, soy products, and beans and other legumes. It is encoded by the codons UUA, UUG, CUU, CUC, CUA, and CUG.
Enoyl-CoA-(∆) isomerase (EC 5.3.3.8, also known as dodecenoyl-CoA- isomerase, 3,2-trans-enoyl-CoA isomerase, ∆3 ,∆2 -enoyl-CoA isomerase, or acetylene-allene isomerase, is an enzyme that catalyzes the conversion of cis- or trans-double bonds of coenzyme A bound fatty acids at gamma-carbon to trans double bonds at beta-carbon as below:
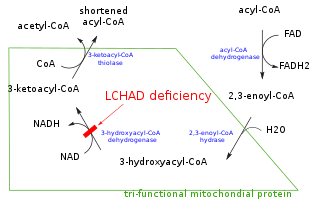
In biochemistry and metabolism, beta-oxidation is the catabolic process by which fatty acid molecules are broken down in the cytosol in prokaryotes and in the mitochondria in eukaryotes to generate acetyl-CoA, which enters the citric acid cycle, and NADH and FADH2, which are co-enzymes used in the electron transport chain. It is named as such because the beta carbon of the fatty acid undergoes oxidation to a carbonyl group. Beta-oxidation is primarily facilitated by the mitochondrial trifunctional protein, an enzyme complex associated with the inner mitochondrial membrane, although very long chain fatty acids are oxidized in peroxisomes.

β-Hydroxy β-methylbutyric acid (HMB), otherwise known as its conjugate base, β-hydroxyβ-methylbutyrate, is a naturally produced substance in humans that is used as a dietary supplement and as an ingredient in certain medical foods that are intended to promote wound healing and provide nutritional support for people with muscle wasting due to cancer or HIV/AIDS. In healthy adults, supplementation with HMB has been shown to increase exercise-induced gains in muscle size, muscle strength, and lean body mass, reduce skeletal muscle damage from exercise, improve aerobic exercise performance, and expedite recovery from exercise. Medical reviews and meta-analyses indicate that HMB supplementation also helps to preserve or increase lean body mass and muscle strength in individuals experiencing age-related muscle loss. HMB produces these effects in part by stimulating the production of proteins and inhibiting the breakdown of proteins in muscle tissue. No adverse effects from long-term use as a dietary supplement in adults have been found.

β-Hydroxybutyric acid, also known as 3-hydroxybutyric acid or BHB, is an organic compound and a beta hydroxy acid with the chemical formula CH3CH(OH)CH2CO2H; its conjugate base is β-hydroxybutyrate, also known as 3-hydroxybutyrate. β-Hydroxybutyric acid is a chiral compound with two enantiomers: D-β-hydroxybutyric acid and L-β-hydroxybutyric acid. Its oxidized and polymeric derivatives occur widely in nature. In humans, D-β-hydroxybutyric acid is one of two primary endogenous agonists of hydroxycarboxylic acid receptor 2 (HCA2), a Gi/o-coupled G protein-coupled receptor (GPCR).

β-Hydroxy β-methylglutaryl-CoA (HMG-CoA), also known as 3-hydroxy-3-methylglutaryl coenzyme A, is an intermediate in the mevalonate and ketogenesis pathways. It is formed from acetyl CoA and acetoacetyl CoA by HMG-CoA synthase. The research of Minor J. Coon and Bimal Kumar Bachhawat in the 1950s at University of Illinois led to its discovery.

Enoyl-CoA hydratase (ECH) or crotonase is an enzyme EC 4.2.1.17 that hydrates the double bond between the second and third carbons on 2-trans/cis-enoyl-CoA:
Methylcrotonyl CoA carboxylase is a biotin-requiring enzyme located in the mitochondria. MCC uses bicarbonate as a carboxyl group source to catalyze the carboxylation of a carbon adjacent to a carbonyl group performing the fourth step in processing leucine, an essential amino acid.

Trifunctional enzyme subunit beta, mitochondrial (TP-beta) also known as 3-ketoacyl-CoA thiolase, acetyl-CoA acyltransferase, or beta-ketothiolase is an enzyme that in humans is encoded by the HADHB gene.
D-Bifunctional protein deficiency is an autosomal recessive peroxisomal fatty acid oxidation disorder. Peroxisomal disorders are usually caused by a combination of peroxisomal assembly defects or by deficiencies of specific peroxisomal enzymes. The peroxisome is an organelle in the cell similar to the lysosome that functions to detoxify the cell. Peroxisomes contain many different enzymes, such as catalase, and their main function is to neutralize free radicals and detoxify drugs. For this reason peroxisomes are ubiquitous in the liver and kidney. D-BP deficiency is the most severe peroxisomal disorder, often resembling Zellweger syndrome.

Isovaleryl-coenzyme A, also known as isovaleryl-CoA, is an intermediate in the metabolism of branched-chain amino acids.

3-Methylcrotonyl-CoA or β-Methylcrotonyl-CoA is an intermediate in the metabolism of leucine.

3-Methylglutaconyl-CoA (MG-CoA), also known as β-methylglutaconyl-CoA, is an intermediate in the metabolism of leucine. It is metabolized into HMG-CoA.
The crotonase family comprises mechanistically diverse proteins that share a conserved trimeric quaternary structure, the core of which consists of 4 turns of a (beta/beta/alpha)n superhelix.

In enzymology, an isovaleryl-CoA dehydrogenase is an enzyme that catalyzes the chemical reaction

In molecular biology, hydroxymethylglutaryl-CoA synthase or HMG-CoA synthase EC 2.3.3.10 is an enzyme which catalyzes the reaction in which acetyl-CoA condenses with acetoacetyl-CoA to form 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA). This reaction comprises the second step in the mevalonate-dependent isoprenoid biosynthesis pathway. HMG-CoA is an intermediate in both cholesterol synthesis and ketogenesis. This reaction is overactivated in patients with diabetes mellitus type 1 if left untreated, due to prolonged insulin deficiency and the exhaustion of substrates for gluconeogenesis and the TCA cycle, notably oxaloacetate. This results in shunting of excess acetyl-CoA into the ketone synthesis pathway via HMG-CoA, leading to the development of diabetic ketoacidosis.

Enoyl Coenzyme A hydratase, short chain, 1, mitochondrial, also known as ECHS1, is a human gene.

α-Ketoisocaproic acid (α-KIC) and its conjugate base, α-ketoisocaproate, are metabolic intermediates in the metabolic pathway for L-leucine. Leucine is an essential amino acid, and its degradation is critical for many biological duties. α-KIC is produced in one of the first steps of the pathway by branched-chain amino acid aminotransferase by transferring the amine on L-leucine onto alpha ketoglutarate, and replacing that amine with a ketone. The degradation of L-leucine in the muscle to this compound allows for the production of the amino acids alanine and glutamate as well. In the liver, α-KIC can be converted to a vast number of compounds depending on the enzymes and cofactors present, including cholesterol, acetyl-CoA, isovaleryl-CoA, and other biological molecules. Isovaleryl-CoA is the main compound synthesized from ɑ-KIC. α-KIC is a key metabolite present in the urine of people with Maple syrup urine disease, along with other branched-chain amino acids. Derivatives of α-KIC have been studied in humans for their ability to improve physical performance during anaerobic exercise as a supplemental bridge between short-term and long-term exercise supplements. These studies show that α-KIC does not achieve this goal without other ergogenicsupplements present as well. α-KIC has also been observed to reduce skeletal muscle damage after eccentrically biased resistance exercises in people who do not usually perform those exercises.

3-hydroxy-3-methylglutaryl-CoA synthase 2 (mitochondrial) is an enzyme in humans that is encoded by the HMGCS2 gene.

β-Hydroxy β-methylbutyryl-coenzyme A (HMB-CoA), also known as 3-hydroxyisovaleryl-CoA, is a metabolite of L-leucine that is produced in the human body. Its immediate precursors are β-hydroxy β-methylbutyric acid (HMB) and β-methylcrotonoyl-CoA (MC-CoA). It can be metabolized into HMB, MC-CoA, and HMG-CoA in humans.