| Saccharopine Dehydrogenase | |||||||
|---|---|---|---|---|---|---|---|
 Saccharopine dehydrogenase from Magnaporthe grisea | |||||||
| Identifiers | |||||||
| Symbol | Saccharop_dh | ||||||
| Pfam | PF03435 | ||||||
| Pfam clan | CL0063 | ||||||
| InterPro | IPR005097 | ||||||
| SCOP2 | 1ff9 / SCOPe / SUPFAM | ||||||
| |||||||
| saccharopine dehydrogenase (putative) | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Symbol | SCCPDH | ||||||
| NCBI gene | 51097 | ||||||
| HGNC | 24275 | ||||||
| RefSeq | NM_016002 | ||||||
| UniProt | Q8NBX0 | ||||||
| Other data | |||||||
| Locus | Chr. 1 q44 | ||||||
| |||||||
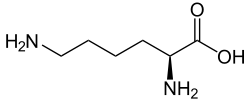
In molecular biology, the protein domain Saccharopine dehydrogenase (SDH), also named Saccharopine reductase, is an enzyme involved in the metabolism of the amino acid lysine, via an intermediate substance called saccharopine. The Saccharopine dehydrogenase enzyme can be classified under EC 1.5.1.7, EC 1.5.1.8, EC 1.5.1.9, and EC 1.5.1.10. It has an important function in lysine metabolism and catalyses a reaction in the α-aminoadipate pathway. This pathway is unique to fungal organisms therefore, this molecule could be useful in the search for new antibiotics. This protein family also includes saccharopine dehydrogenase and homospermidine synthase. It is found in prokaryotes, eukaryotes and archaea.