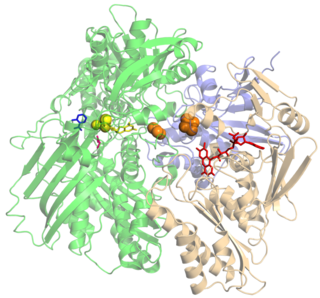
Xanthine oxidase is a form of xanthine oxidoreductase, a type of enzyme that generates reactive oxygen species. These enzymes catalyze the oxidation of hypoxanthine to xanthine and can further catalyze the oxidation of xanthine to uric acid. These enzymes play an important role in the catabolism of purines in some species, including humans.
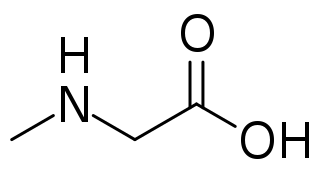
Sarcosine, also known as N-methylglycine, or monomethylglycine, is a amino acid with the formula CH3N(H)CH2CO2H. It exists at neutral pH as the zwitterion CH3N+(H)2CH2CO2−, which can be obtained as a white, water-soluble powder. Like some amino acids, sarcosine converts to a cation at low pH and an anion at high pH, with the respective formulas CH3N+(H)2CH2CO2H and CH3N(H)CH2CO2−. Sarcosine is a close relative of glycine, with a secondary amine in place of the primary amine.

The glucose oxidase enzyme also known as notatin is an oxidoreductase that catalyses the oxidation of glucose to hydrogen peroxide and D-glucono-δ-lactone. This enzyme is produced by certain species of fungi and insects and displays antibacterial activity when oxygen and glucose are present.
Purine metabolism refers to the metabolic pathways to synthesize and break down purines that are present in many organisms.

10-Formyltetrahydrofolate (10-CHO-THF) is a form of tetrahydrofolate that acts as a donor of formyl groups in anabolism. In these reactions 10-CHO-THF is used as a substrate in formyltransferase reactions.
Glutamate formimidoyltransferase is a methyltransferase enzyme which uses tetrahydrofolate as part of histidine catabolism. It catalyses two reactions:
In enzymology, sarcosine dehydrogenase (EC 1.5.8.3) is a mitochondrial enzyme that catalyzes the chemical reaction N-demethylation of sarcosine to give glycine. This enzyme belongs to the family of oxidoreductases, specifically those acting on the CH-NH group of donor with other acceptors. The systematic name of this enzyme class is sarcosine:acceptor oxidoreductase (demethylating). Other names in common use include sarcosine N-demethylase, monomethylglycine dehydrogenase, and sarcosine:(acceptor) oxidoreductase (demethylating). Sarcosine dehydrogenase is closely related to dimethylglycine dehydrogenase, which catalyzes the demethylation reaction of dimethylglycine to sarcosine. Both sarcosine dehydrogenase and dimethylglycine dehydrogenase use FAD as a cofactor. Sarcosine dehydrogenase is linked by electron-transferring flavoprotein (ETF) to the respiratory redox chain. The general chemical reaction catalyzed by sarcosine dehydrogenase is:
In enzymology, a trimethylsulfonium-tetrahydrofolate N-methyltransferase is an enzyme that catalyzes the chemical reaction

In enzymology, an alcohol oxidase (EC 1.1.3.13) is an enzyme that catalyzes the chemical reaction
In enzymology, a dimethylglycine dehydrogenase (EC 1.5.8.4) is an enzyme that catalyzes the chemical reaction
In enzymology, a dimethylglycine oxidase (EC 1.5.3.10) is an enzyme that catalyzes the chemical reaction
In enzymology, a 3-methyl-2-oxobutanoate hydroxymethyltransferase (EC 2.1.2.11) is an enzyme that catalyzes the chemical reaction

In enzymology, a formate—tetrahydrofolate ligase is an enzyme that catalyzes the chemical reaction

FAD-dependent oxidoreductase domain-containing protein 1 (FOXRED1), also known as H17, or FP634 is an enzyme that in humans is encoded by the FOXRED1 gene. FOXRED1 is an oxidoreductase and complex I-specific molecular chaperone involved in the assembly and stabilization of NADH dehydrogenase (ubiquinone) also known as complex I, which is located in the mitochondrial inner membrane and is the largest of the five complexes of the electron transport chain. Mutations in FOXRED1 have been associated with Leigh syndrome and infantile-onset mitochondrial encephalopathy.

In molecular biology, the FAD dependent oxidoreductase family of proteins is a family of FAD dependent oxidoreductases. Members of this family include Glycerol-3-phosphate dehydrogenase EC 1.1.99.5, Sarcosine oxidase beta subunit EC 1.5.3.1, D-amino-acid dehydrogenase EC 1.4.99.1, D-aspartate oxidase EC 1.4.3.1.
Glycine/sarcosine N-methyltransferase is an enzyme with systematic name S-adenosyl-L-methionine:glycine(or sarcosine) N-methyltransferase . This enzyme catalyses the following chemical reaction
Sarcosine/dimethylglycine N-methyltransferase is an enzyme with systematic name S-adenosyl-L-methionine:sarcosine(or N,N-dimethylglycine) N-methyltransferase . This enzyme catalyses the following chemical reaction
Glycine/sarcosine/dimethylglycine N-methyltransferase is an enzyme with systematic name S-adenosyl-L-methionine:glycine(or sarcosine or N,N-dimethylglycine) N-methyltransferase . This enzyme catalyses the following chemical reaction
In organic chemistry, secondary amino acids are amino acids which do not contain the amino group −NH2 but is rather a secondary amine. Secondary amino acids can be classified to cyclic acids, such as proline, and acyclic N-substituted amino acids.
This page is based on this
Wikipedia article Text is available under the
CC BY-SA 4.0 license; additional terms may apply.
Images, videos and audio are available under their respective licenses.








