
Histidine (symbol His or H) is an essential α-amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated –NH3+ form under biological conditions), a carboxylic acid group (which is in the deprotonated –COO− form under biological conditions), and an imidazole side chain (which is partially protonated), classifying it as a positively charged amino acid at physiological pH. Initially thought essential only for infants, it has now been shown in longer-term studies to be essential for adults also. It is encoded by the codons CAU and CAC.

Malate dehydrogenase (EC 1.1.1.37) (MDH) is an enzyme that reversibly catalyzes the oxidation of malate to oxaloacetate using the reduction of NAD+ to NADH. This reaction is part of many metabolic pathways, including the citric acid cycle. Other malate dehydrogenases, which have other EC numbers and catalyze other reactions oxidizing malate, have qualified names like malate dehydrogenase (NADP+).

Aconitase is an enzyme that catalyses the stereo-specific isomerization of citrate to isocitrate via cis-aconitate in the tricarboxylic acid cycle, a non-redox-active process.

Lipoxygenases are a family of (non-heme) iron-containing enzymes most of which catalyze the dioxygenation of polyunsaturated fatty acids in lipids containing a cis,cis-1,4- pentadiene into cell signaling agents that serve diverse roles as autocrine signals that regulate the function of their parent cells, paracrine signals that regulate the function of nearby cells, and endocrine signals that regulate the function of distant cells.
The branched-chain α-ketoacid dehydrogenase complex is a multi-subunit complex of enzymes that is found on the mitochondrial inner membrane. This enzyme complex catalyzes the oxidative decarboxylation of branched, short-chain alpha-ketoacids. BCKDC is a member of the mitochondrial α-ketoacid dehydrogenase complex family comprising pyruvate dehydrogenase and alpha-ketoglutarate dehydrogenase, key enzymes that function in the Krebs cycle.

Methylmalonyl-CoA mutase (EC 5.4.99.2, MCM), mitochondrial, also known as methylmalonyl-CoA isomerase, is a protein that in humans is encoded by the MUT gene. This vitamin B12-dependent enzyme catalyzes the isomerization of methylmalonyl-CoA to succinyl-CoA in humans. Mutations in MUT gene may lead to various types of methylmalonic aciduria.

Urocanic acid is an intermediate in the catabolism of L-histidine.
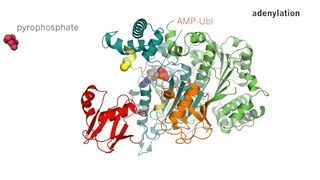
Enzyme catalysis is the increase in the rate of a process by a biological molecule, an "enzyme". Most enzymes are proteins, and most such processes are chemical reactions. Within the enzyme, generally catalysis occurs at a localized site, called the active site.

6-Phosphogluconolactonase is a cytosolic enzyme found in all organisms that catalyzes the hydrolysis of 6-phosphogluconolactone to 6-phosphogluconic acid in the oxidative phase of the pentose phosphate pathway. The tertiary structure of 6PGL employs an α/β hydrolase fold, with active site residues clustered on the loops of the α-helices. Based on the crystal structure of the enzyme, the mechanism is proposed to be dependent on proton transfer by a histidine residue in the active site. 6PGL selectively catalyzes the hydrolysis of δ-6-phosphogluconolactone, and has no activity on the γ isomer.

Methylmalonyl CoA epimerase is an enzyme involved in fatty acid catabolism that is encoded in human by the "MCEE" gene located on chromosome 2. It is routinely and incorrectly labeled as "methylmalonyl-CoA racemase". It is not a racemase because the CoA moiety has 5 other stereocenters.

Histidine ammonia-lyase is an enzyme that in humans is encoded by the HAL gene. Histidase converts histidine into ammonia and urocanic acid.

3-Methylglutaconyl-CoA hydratase, also known as MG-CoA hydratase and AUH, is an enzyme encoded by the AUH gene on chromosome 19. It is a member of the enoyl-CoA hydratase/isomerase superfamily, but it is the only member of that family that is able to bind to RNA. Not only does it bind to RNA, AUH has also been observed to be involved in the metabolic enzymatic activity, making it a dual-role protein. Mutations of this gene have been found to cause a disease called 3-Methylglutaconic Acuduria Type 1.

In enzymology, a homoserine dehydrogenase (EC 1.1.1.3) is an enzyme that catalyzes the chemical reaction
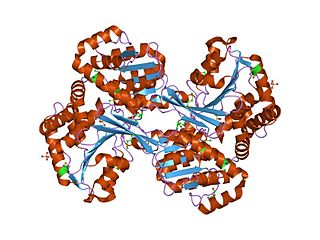
In enzymology, a histidinol dehydrogenase (HIS4) (HDH) (EC 1.1.1.23) is an enzyme that catalyzes the chemical reaction
In enzymology, a 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase (EC 1.3.1.28) is an enzyme that catalyzes the chemical reaction

In enzymology, an acyl-[acyl-carrier-protein] desaturase (EC 1.14.19.2) is an enzyme that catalyzes the chemical reaction

In enzymology, a microsomal epoxide hydrolase (mEH) is an enzyme that catalyzes the hydrolysis reaction between an epoxide and water to form a diol.
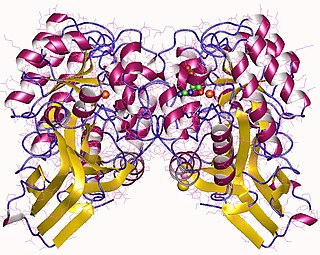
In enzymology, an imidazolonepropionase (EC 3.5.2.7) is an enzyme that catalyzes the chemical reaction

Urocanic aciduria is an autosomal recessive metabolic disorder caused by a deficiency of the enzyme urocanase. It is a secondary disorder of histidine metabolism.
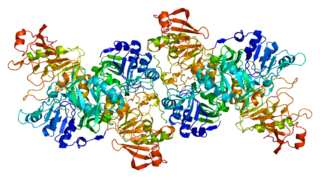
Coenzyme A transferases (CoA-transferases) are transferase enzymes that catalyze the transfer of a coenzyme A group from an acyl-CoA donor to a carboxylic acid acceptor. Among other roles, they are responsible for transfer of CoA groups during fermentation and metabolism of ketone bodies. These enzymes are found in all three domains of life.



















