A protein phosphatase is a phosphatase enzyme that removes a phosphate group from the phosphorylated amino acid residue of its substrate protein. Protein phosphorylation is one of the most common forms of reversible protein posttranslational modification (PTM), with up to 30% of all proteins being phosphorylated at any given time. Protein kinases (PKs) are the effectors of phosphorylation and catalyse the transfer of a γ-phosphate from ATP to specific amino acids on proteins. Several hundred PKs exist in mammals and are classified into distinct super-families. Proteins are phosphorylated predominantly on Ser, Thr and Tyr residues, which account for 79.3, 16.9 and 3.8% respectively of the phosphoproteome, at least in mammals. In contrast, protein phosphatases (PPs) are the primary effectors of dephosphorylation and can be grouped into three main classes based on sequence, structure and catalytic function. The largest class of PPs is the phosphoprotein phosphatase (PPP) family comprising PP1, PP2A, PP2B, PP4, PP5, PP6 and PP7, and the protein phosphatase Mg2+- or Mn2+-dependent (PPM) family, composed primarily of PP2C. The protein Tyr phosphatase (PTP) super-family forms the second group, and the aspartate-based protein phosphatases the third. The protein pseudophosphatases form part of the larger phosphatase family, and in most cases are thought to be catalytically inert, instead functioning as phosphate-binding proteins, integrators of signalling or subcellular traps. Examples of membrane-spanning protein phosphatases containing both active (phosphatase) and inactive (pseudophosphatase) domains linked in tandem are known, conceptually similar to the kinase and pseudokinase domain polypeptide structure of the JAK pseudokinases. A complete comparative analysis of human phosphatases and pseudophosphatases has been completed by Manning and colleagues, forming a companion piece to the ground-breaking analysis of the human kinome, which encodes the complete set of ~536 human protein kinases.

In biochemistry, a kinase is an enzyme that catalyzes the transfer of phosphate groups from high-energy, phosphate-donating molecules to specific substrates. This process is known as phosphorylation, where the high-energy ATP molecule donates a phosphate group to the substrate molecule. As a result, kinase produces a phosphorylated substrate and ADP. Conversely, it is referred to as dephosphorylation when the phosphorylated substrate donates a phosphate group and ADP gains a phosphate group. These two processes, phosphorylation and dephosphorylation, occur four times during glycolysis.

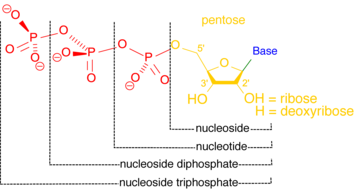
Adenosine monophosphate (AMP), also known as 5'-adenylic acid, is a nucleotide. AMP consists of a phosphate group, the sugar ribose, and the nucleobase adenine. It is an ester of phosphoric acid and the nucleoside adenosine. As a substituent it takes the form of the prefix adenylyl-.
Gluconeogenesis (GNG) is a metabolic pathway that results in the biosynthesis of glucose from certain non-carbohydrate carbon substrates. It is a ubiquitous process, present in plants, animals, fungi, bacteria, and other microorganisms. In vertebrates, gluconeogenesis occurs mainly in the liver and, to a lesser extent, in the cortex of the kidneys. It is one of two primary mechanisms – the other being degradation of glycogen (glycogenolysis) – used by humans and many other animals to maintain blood sugar levels, avoiding low levels (hypoglycemia). In ruminants, because dietary carbohydrates tend to be metabolized by rumen organisms, gluconeogenesis occurs regardless of fasting, low-carbohydrate diets, exercise, etc. In many other animals, the process occurs during periods of fasting, starvation, low-carbohydrate diets, or intense exercise.

Guanosine-5'-triphosphate (GTP) is a purine nucleoside triphosphate. It is one of the building blocks needed for the synthesis of RNA during the transcription process. Its structure is similar to that of the guanosine nucleoside, the only difference being that nucleotides like GTP have phosphates on their ribose sugar. GTP has the guanine nucleobase attached to the 1' carbon of the ribose and it has the triphosphate moiety attached to ribose's 5' carbon.

Guanosine diphosphate, abbreviated GDP, is a nucleoside diphosphate. It is an ester of pyrophosphoric acid with the nucleoside guanosine. GDP consists of a pyrophosphate group, a pentose sugar ribose, and the nucleobase guanine.
Adenylate kinase is a phosphotransferase enzyme that catalyzes the interconversion of the various adenosine phosphates. By constantly monitoring phosphate nucleotide levels inside the cell, ADK plays an important role in cellular energy homeostasis.
In biochemistry, dephosphorylation is the removal of a phosphate group from an organic compound by hydrolysis. It is a reversible post-translational modification. Dephosphorylation and its counterpart, phosphorylation, activate and deactivate enzymes by detaching or attaching phosphoric esters and anhydrides. A notable occurrence of dephosphorylation is the conversion of ATP to ADP and inorganic phosphate.
A regulatory enzyme is an enzyme in a biochemical pathway which, through its responses to the presence of certain other biomolecules, regulates the pathway activity. This is usually done for pathways whose products may be needed in different amounts at different times, such as hormone production. Regulatory enzymes exist at high concentrations so their activity can be increased or decreased with changes in substrate concentrations

Nucleoside-diphosphate kinases are enzymes that catalyze the exchange of terminal phosphate between different nucleoside diphosphates (NDP) and triphosphates (NTP) in a reversible manner to produce nucleotide triphosphates. Many NDP serve as acceptor while NTP are donors of phosphate group. The general reaction via ping-pong mechanism is as follows: XDP + YTP ←→ XTP + YDP. NDPK activities maintain an equilibrium between the concentrations of different nucleoside triphosphates such as, for example, when guanosine triphosphate (GTP) produced in the citric acid (Krebs) cycle is converted to adenosine triphosphate (ATP). Other activities include cell proliferation, differentiation and development, signal transduction, G protein-coupled receptor, endocytosis, and gene expression.
Phosphofructokinase-2 (6-phosphofructo-2-kinase, PFK-2) or fructose bisphosphatase-2 (FBPase-2), is an enzyme indirectly responsible for regulating the rates of glycolysis and gluconeogenesis in cells. It catalyzes formation and degradation of a significant allosteric regulator, fructose-2,6-bisphosphate (Fru-2,6-P2) from substrate fructose-6-phosphate. Fru-2,6-P2 contributes to the rate-determining step of glycolysis as it activates enzyme phosphofructokinase 1 in the glycolysis pathway, and inhibits fructose-1,6-bisphosphatase 1 in gluconeogenesis. Since Fru-2,6-P2 differentially regulates glycolysis and gluconeogenesis, it can act as a key signal to switch between the opposing pathways. Because PFK-2 produces Fru-2,6-P2 in response to hormonal signaling, metabolism can be more sensitively and efficiently controlled to align with the organism's glycolytic needs. This enzyme participates in fructose and mannose metabolism. The enzyme is important in the regulation of hepatic carbohydrate metabolism and is found in greatest quantities in the liver, kidney and heart. In mammals, several genes often encode different isoforms, each of which differs in its tissue distribution and enzymatic activity. The family described here bears a resemblance to the ATP-driven phospho-fructokinases; however, they share little sequence similarity, although a few residues seem key to their interaction with fructose 6-phosphate.

5′-Nucleotidase is an enzyme which catalyzes the phosphorylytic cleavage of 5′-nucleotides. Although originally found in snake venom, the activity of 5'nucleotidase has been described for bacteria and plant cells, and is widely distributed in vertebrate tissue. In mammalian cells the enzyme is predominantly located in the plasma membrane and its primary role is in the conversion of extracellular nucleotides, which are generally impermeable, to the corresponding nucleoside which can readily enter most cells. Consequently, the enzyme plays a key role in the metabolism of nucleotides.
A nucleotidase is a hydrolytic enzyme that catalyzes the hydrolysis of a nucleotide into a nucleoside and a phosphate.

Fructose 2,6-bisphosphate, abbreviated Fru-2,6-P2, is a metabolite that allosterically affects the activity of the enzymes phosphofructokinase 1 (PFK-1) and fructose 1,6-bisphosphatase (FBPase-1) to regulate glycolysis and gluconeogenesis. Fru-2,6-P2 itself is synthesized and broken down in either direction by the integrated bifunctional enzyme phosphofructokinase 2 (PFK-2/FBPase-2), which also contains a phosphatase domain and is also known as fructose-2,6-bisphosphatase. Whether the kinase and phosphatase domains of PFK-2/FBPase-2 are active or inactive depends on the phosphorylation state of the enzyme.

Serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform is an enzyme that is encoded by the PPP2CA gene.

The enzyme phosphatidate phosphatase (PAP, EC 3.1.3.4) is a key regulatory enzyme in lipid metabolism, catalyzing the conversion of phosphatidate to diacylglycerol:
In enzymology, a nucleoside-phosphate kinase is an enzyme that catalyzes the chemical reaction

5', 3'-nucleotidase, cytosolic, also known as 5'(3')-deoxyribonucleotidase, cytosolic type (cdN) or deoxy-5'-nucleotidase 1 (dNT-1), is an enzyme that in humans is encoded by the NT5C gene on chromosome 17.

5',3'-nucleotidase, mitochondrial, also known as 5'(3')-deoxyribonucleotidase, mitochondrial (mdN) or deoxy-5'-nucleotidase 2 (dNT-2), is an enzyme that in humans is encoded by the NT5M gene. This gene encodes a 5' nucleotidase that localizes to the mitochondrial matrix. This enzyme dephosphorylates the 5'- and 2'(3')-phosphates of uracil and thymine deoxyribonucleotides. The gene is located within the Smith–Magenis syndrome region on chromosome 17.
The phosphatome of an organism is the set of phosphatase genes in its genome. Phosphatases are enzymes that catalyze the removal of phosphate from biomolecules. Over half of all cellular proteins are modified by phosphorylation which typically controls their functions. Protein phosphorylation is controlled by the opposing actions of protein phosphatases and protein kinases. Most phosphorylation sites are not linked to a specific phosphatase, so the phosphatome approach allows a global analysis of dephosphorylation, screening to find the phosphatase responsible for a given reaction, and comparative studies between different phosphatases, similar to how protein kinase research has been impacted by the kinome approach.