In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part of biological inheritance. This is essential for cell division during growth and repair of damaged tissues, while it also ensures that each of the new cells receives its own copy of the DNA. The cell possesses the distinctive property of division, which makes replication of DNA essential.

A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create two identical DNA duplexes from a single original DNA duplex. During this process, DNA polymerase "reads" the existing DNA strands to create two new strands that match the existing ones. These enzymes catalyze the chemical reaction
DNA primase is an enzyme involved in the replication of DNA and is a type of RNA polymerase. Primase catalyzes the synthesis of a short RNA segment called a primer complementary to a ssDNA template. After this elongation, the RNA piece is removed by a 5' to 3' exonuclease and refilled with DNA.

Helicases are a class of enzymes thought to be vital to all organisms. Their main function is to unpack an organism's genetic material. Helicases are motor proteins that move directionally along a nucleic acid phosphodiester backbone, separating two hybridized nucleic acid strands, using energy from ATP hydrolysis. There are many helicases, representing the great variety of processes in which strand separation must be catalyzed. Approximately 1% of eukaryotic genes code for helicases.

In biochemistry, a nuclease is an enzyme capable of cleaving the phosphodiester bonds that link nucleotides together to form nucleic acids. Nucleases variously affect single and double stranded breaks in their target molecules. In living organisms, they are essential machinery for many aspects of DNA repair. Defects in certain nucleases can cause genetic instability or immunodeficiency. Nucleases are also extensively used in molecular cloning.

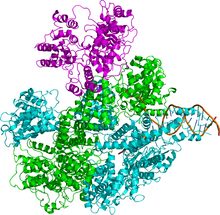
RuvABC is a complex of three proteins that mediate branch migration and resolve the Holliday junction created during homologous recombination in bacteria. As such, RuvABC is critical to bacterial DNA repair.
DNA gyrase, or simply gyrase, is an enzyme within the class of topoisomerase and is a subclass of Type II topoisomerases that reduces topological strain in an ATP dependent manner while double-stranded DNA is being unwound by elongating RNA-polymerase or by helicase in front of the progressing replication fork. It is the only known enzyme to actively contribute negative supercoiling to DNA, while it also is capable of relaxing positive supercoils. It does so by looping the template to form a crossing, then cutting one of the double helices and passing the other through it before releasing the break, changing the linking number by two in each enzymatic step. This process occurs in bacteria, whose single circular DNA is cut by DNA gyrase and the two ends are then twisted around each other to form supercoils. Gyrase is also found in eukaryotic plastids: it has been found in the apicoplast of the malarial parasite Plasmodium falciparum and in chloroplasts of several plants. Bacterial DNA gyrase is the target of many antibiotics, including nalidixic acid, novobiocin, albicidin, and ciprofloxacin.
RecQ helicase is a family of helicase enzymes initially found in Escherichia coli that has been shown to be important in genome maintenance. They function through catalyzing the reaction ATP + H2O → ADP + P and thus driving the unwinding of paired DNA and translocating in the 3' to 5' direction. These enzymes can also drive the reaction NTP + H2O → NDP + P to drive the unwinding of either DNA or RNA.

Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolyzing reaction that breaks phosphodiester bonds at either the 3′ or the 5′ end occurs. Its close relative is the endonuclease, which cleaves phosphodiester bonds in the middle (endo) of a polynucleotide chain. Eukaryotes and prokaryotes have three types of exonucleases involved in the normal turnover of mRNA: 5′ to 3′ exonuclease (Xrn1), which is a dependent decapping protein; 3′ to 5′ exonuclease, an independent protein; and poly(A)-specific 3′ to 5′ exonuclease.

Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids.

Werner syndrome ATP-dependent helicase, also known as DNA helicase, RecQ-like type 3, is an enzyme that in humans is encoded by the WRN gene. WRN is a member of the RecQ Helicase family. Helicase enzymes generally unwind and separate double-stranded DNA. These activities are necessary before DNA can be copied in preparation for cell division. Helicase enzymes are also critical for making a blueprint of a gene for protein production, a process called transcription. Further evidence suggests that Werner protein plays a critical role in repairing DNA. Overall, this protein helps maintain the structure and integrity of a person's DNA.
Deoxyribonuclease IV (phage-T4-induced) is catalyzes the degradation nucleotides in DsDNA by attacking the 5'-terminal end.

Exonuclease 1 is an enzyme that in humans is encoded by the EXO1 gene.

ATP-dependent DNA helicase Q1 is an enzyme that in humans is encoded by the RECQL gene.

T7 DNA polymerase is an enzyme used during the DNA replication of the T7 bacteriophage. During this process, the DNA polymerase “reads” existing DNA strands and creates two new strands that match the existing ones. The T7 DNA polymerase requires a host factor, E. coli thioredoxin, in order to carry out its function. This helps stabilize the binding of the necessary protein to the primer-template to improve processivity by more than 100-fold, which is a feature unique to this enzyme. It is a member of the Family A DNA polymerases, which include E. coli DNA polymerase I and Taq DNA polymerase.
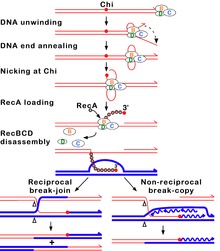
A Chi site or Chi sequence is a short stretch of DNA in the genome of a bacterium near which homologous recombination is more likely to occur than on average across the genome. Chi sites serve as stimulators of DNA double-strand break repair in bacteria, which can arise from radiation or chemical treatments, or result from replication fork breakage during DNA replication. The sequence of the Chi site is unique to each group of closely related organisms; in E. coli and other enteric bacteria, such as Salmonella, the core sequence is 5'-GCTGGTGG-3' plus important nucleotides about 4 to 7 nucleotides to the 3' side of the core sequence. The existence of Chi sites was originally discovered in the genome of bacteriophage lambda, a virus that infects E. coli, but is now known to occur about 1000 times in the E. coli genome.

A circular chromosome is a chromosome in bacteria, archaea, mitochondria, and chloroplasts, in the form of a molecule of circular DNA, unlike the linear chromosome of most eukaryotes.
The RecF pathway, also called the RecFOR pathway, is a pathway of homologous recombination that repairs DNA in bacteria. It repairs breaks that occur on only one of DNA's two strands, known as single-strand gaps. The RecF pathway can also repair double-strand breaks in DNA when the RecBCD pathway, another pathway of homologous recombination in bacteria, is inactivated by mutations. Like the RecBCD pathway, the RecF pathway requires RecA for strand invasion. The two pathways are also similar in their phases of branch migration, in which the Holliday junction slides in one direction, and resolution, in which the Holliday junctions are cleaved apart by enzymes.
DNA Polymerase V is a polymerase enzyme involved in DNA repair mechanisms in bacteria, such as Escherichia coli. It is composed of a UmuD' homodimer and a UmuC monomer, forming the UmuD'2C protein complex. It is part of the Y-family of DNA Polymerases, which are capable of performing DNA translesion synthesis (TLS). Translesion polymerases bypass DNA damage lesions during DNA replication - if a lesion is not repaired or bypassed the replication fork can stall and lead to cell death. However, Y polymerases have low sequence fidelity during replication. When the UmuC and UmuD' proteins were initially discovered in E. coli, they were thought to be agents that inhibit faithful DNA replication and caused DNA synthesis to have high mutation rates after exposure to UV-light. The polymerase function of Pol V was not discovered until the late 1990s when UmuC was successfully extracted, consequent experiments unequivocally proved UmuD'2C is a polymerase. This finding lead to the detection of many Pol V orthologs and the discovery of the Y-family of polymerases.
Stephen Charles Kowalczykowski is a Distinguished Professor of Microbiology and Molecular Genetics at the University of California at Davis. His research focuses on the biochemistry and molecular biology of DNA repair and homologous recombination. His lab combines fluorescence microscopy, optical trapping and microfluidics to manipulate and visualize single molecules of DNA and the enzymes involved in processing and repairing DNA. He calls this scientific approach, "visual biochemistry". Stephen Kowalczykowski was elected to the American Society for Arts and Science in 2005, the National Academy of Sciences in 2007 and was a Harvey Society Lecturer at Rockefeller University in 2012.