Related Research Articles
A cistron is a region of DNA that is conceptually equivalent to some definitions of a gene, such that the terms are synonymous from certain viewpoints, especially with regard to the molecular gene as contrasted with the Mendelian gene. The question of which scope of a subset of DNA constitutes a unit of selection is the question that governs whether cistrons are the same thing as genes. The word cistron is used to emphasize that molecular genes exhibit a specific behavior in a complementation test ; distinct positions within a genome are cistronic.
Genetic linkage is the tendency of DNA sequences that are close together on a chromosome to be inherited together during the meiosis phase of sexual reproduction. Two genetic markers that are physically near to each other are unlikely to be separated onto different chromatids during chromosomal crossover, and are therefore said to be more linked than markers that are far apart. In other words, the nearer two genes are on a chromosome, the lower the chance of recombination between them, and the more likely they are to be inherited together. Markers on different chromosomes are perfectly unlinked, although the penetrance of potentially deleterious alleles may be influenced by the presence of other alleles, and these other alleles may be located on other chromosomes than that on which a particular potentially deleterious allele is located.

Molecular genetics is a branch of biology that addresses how differences in the structures or expression of DNA molecules manifests as variation among organisms. Molecular genetics often applies an "investigative approach" to determine the structure and/or function of genes in an organism's genome using genetic screens.
The Crick, Brenner et al. experiment (1961) was a scientific experiment performed by Francis Crick, Sydney Brenner, Leslie Barnett and R.J. Watts-Tobin. It was a key experiment in the development of what is now known as molecular biology and led to a publication entitled "The General Nature of the Genetic Code for Proteins" and according to the historian of Science Horace Judson is "regarded...as a classic of intellectual clarity, precision and rigour". This study demonstrated that the genetic code is made up of a series of three base pair codons which code for individual amino acids. The experiment also elucidated the nature of gene expression and frame-shift mutations.

Escherichia virus T4 is a species of bacteriophages that infect Escherichia coli bacteria. It is a double-stranded DNA virus in the subfamily Tevenvirinae of the family Straboviridae. T4 is capable of undergoing only a lytic life cycle and not the lysogenic life cycle. The species was formerly named T-even bacteriophage, a name which also encompasses, among other strains, Enterobacteria phage T2, Enterobacteria phage T4 and Enterobacteria phage T6.

Seymour Benzer was an American physicist, molecular biologist and behavioral geneticist. His career began during the molecular biology revolution of the 1950s, and he eventually rose to prominence in the fields of molecular and behavioral genetics. He led a productive genetics research lab both at Purdue University and as the James G. Boswell Professor of Neuroscience, emeritus, at the California Institute of Technology.

Franklin (Frank) William Stahl is an American molecular biologist and geneticist. With Matthew Meselson, Stahl conducted the famous Meselson-Stahl experiment showing that DNA is replicated by a semiconservative mechanism, meaning that each strand of the DNA serves as a template for production of a new strand.
Complementation refers to a genetic process when two strains of an organism with different homozygous recessive mutations that produce the same mutant phenotype have offspring that express the wild-type phenotype when mated or crossed. Complementation will ordinarily occur if the mutations are in different genes. Complementation may also occur if the two mutations are at different sites within the same gene, but this effect is usually weaker than that of intergenic complementation. When the mutations are in different genes, each strain's genome supplies the wild-type allele to "complement" the mutated allele of the other strain's genome. Since the mutations are recessive, the offspring will display the wild-type phenotype. A complementation test can test whether the mutations in two strains are in different genes. Complementation is usually weaker or absent if the mutations are in the same gene. The convenience and essence of this test is that the mutations that produce a phenotype can be assigned to different genes without the exact knowledge of what the gene product is doing on a molecular level. American geneticist Edward B. Lewis developed the complementation test.
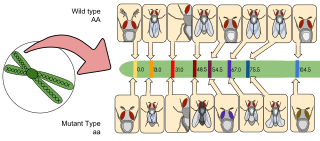
Gene mapping or genome mapping describes the methods used to identify the location of a gene on a chromosome and the distances between genes. Gene mapping can also describe the distances between different sites within a gene.
A suppressor mutation is a second mutation that alleviates or reverts the phenotypic effects of an already existing mutation in a process defined synthetic rescue. Genetic suppression therefore restores the phenotype seen prior to the original background mutation. Suppressor mutations are useful for identifying new genetic sites which affect a biological process of interest. They also provide evidence between functionally interacting molecules and intersecting biological pathways.
A nonsense suppressor is a factor which can inhibit the effect of the nonsense mutation. Nonsense suppressors can be generally divided into two classes: a) a mutated tRNA which can bind with a termination codon on mRNA; b) a mutation on ribosomes decreasing the effect of a termination codon. It is believed that nonsense suppressors keep a low concentration in the cell and do not disrupt normal translation most of the time. In addition, many genes do not have only one termination codon, and cells commonly use ochre codons as the termination signal, whose nonsense suppressors are usually inefficient.
Temperature-sensitive mutants are variants of genes that allow normal function of the organism at low temperatures, but altered function at higher temperatures. Cold sensitive mutants are variants of genes that allow normal function of the organism at higher temperatures, but altered function at low temperatures.
Lethal alleles are alleles that cause the death of the organism that carries them. They are usually a result of mutations in genes that are essential for growth or development. Lethal alleles may be recessive, dominant, or conditional depending on the gene or genes involved.
The phage group was an informal network of biologists centered on Max Delbrück that contributed heavily to bacterial genetics and the origins of molecular biology in the mid-20th century. The phage group takes its name from bacteriophages, the bacteria-infecting viruses that the group used as experimental model organisms. In addition to Delbrück, important scientists associated with the phage group include: Salvador Luria, Alfred Hershey, Seymour Benzer, Charles Steinberg, Gunther Stent, James D. Watson, Frank Stahl, and Renato Dulbecco.
The term proofreading is used in genetics to refer to the error-correcting processes, first proposed by John Hopfield and Jacques Ninio, involved in DNA replication, immune system specificity, and enzyme-substrate recognition among many other processes that require enhanced specificity. The proofreading mechanisms of Hopfield and Ninio are non-equilibrium active processes that consume ATP to enhance specificity of various biochemical reactions.
Leslie Barnett was a British biologist who worked with Francis Crick, Sydney Brenner, and Richard J. Watts-Tobin to genetically demonstrate the triplet nature of the code of protein translation through the Crick, Brenner, Barnett, Watts-Tobin et al. experiment of 1961, which discovered frameshift mutations; this insight provided early elucidation of the nature of the genetic code.
Experimental evolution studies are a means of testing evolutionary theory under carefully designed, reproducible experiments. Given enough time, space, and money, any organism could be used for experimental evolution studies. However, those with rapid generation times, high mutation rates, large population sizes, and small sizes increase the feasibility of experimental studies in a laboratory context. For these reasons, bacteriophages are especially favored by experimental evolutionary biologists. Bacteriophages, and microbial organisms, can be frozen in stasis, facilitating comparison of evolved strains to ancestors. Additionally, microbes are especially labile from a molecular biologic perspective. Many molecular tools have been developed to manipulate the genetic material of microbial organisms, and because of their small genome sizes, sequencing the full genomes of evolved strains is trivial. Therefore, comparisons can be made for the exact molecular changes in evolved strains during adaptation to novel conditions.
The history of genetics can be represented on a timeline of events from the earliest work in the 1850s, to the DNA era starting in the 1940s, and the genomics era beginning in the 1970s.

Epistasis is a phenomenon in genetics in which the effect of a gene mutation is dependent on the presence or absence of mutations in one or more other genes, respectively termed modifier genes. In other words, the effect of the mutation is dependent on the genetic background in which it appears. Epistatic mutations therefore have different effects on their own than when they occur together. Originally, the term epistasis specifically meant that the effect of a gene variant is masked by that of different gene.
Charles 'Charley' M. Steinberg was an immunobiologist and permanent member of the Basel Institute for Immunology. He was a former student of Max Delbrück. Notably he hosted Richard Feynman at Caltech when Feynman studied molecular biology, leading Feynman to remark that Charlie was “...the smartest guy I know”. He was instrumental in the discovery of V(D)J recombination, bacteriophage genetics as part of the phage group and co-discoverer of the amber-mutant of the T4 bacteriophage that led to the recognition of stop codons.
References
- R Jayaraman. "Seymour Benzer and T4 rII: Running the Map into the Ground." Resonance, October 2008, pp. 898–908.
- Jonathan Weiner. Time, Love, Memory: A Great Biologist and His Quest for the Origins of Behavior. Knopf. ISBN 0-679-44435-1