Related Research Articles

In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part for biological inheritance. The cell possesses the distinctive property of division, which makes replication of DNA essential.
Enterobacteria phage λ is a bacterial virus, or bacteriophage, that infects the bacterial species Escherichia coli. It was discovered by Esther Lederberg in 1950. The wild type of this virus has a temperate life cycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase, during which it kills and lyses the cell to produce offspring. Lambda strains, mutated at specific sites, are unable to lysogenize cells; instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell.

DNA polymerase is an enzyme that synthesizes DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create two identical DNA duplexes from a single original DNA duplex. During this process, DNA polymerase "reads" the existing DNA strands to create two new strands that match the existing ones. These enzymes catalyze the chemical reaction

A nuclease is an enzyme capable of cleaving the phosphodiester bonds between nucleotides of nucleic acids. Nucleases variously effect single and double stranded breaks in their target molecules. In living organisms, they are essential machinery for many aspects of DNA repair. Defects in certain nucleases can cause genetic instability or immunodeficiency. Nucleases are also extensively used in molecular cloning.

Okazaki fragments are short sequences of DNA nucleotides which are synthesized discontinuously and later linked together by the enzyme DNA ligase to create the lagging strand during DNA replication. They were discovered in the 1960s by the Japanese molecular biologists Reiji and Tsuneko Okazaki, along with the help of some of their colleagues.

RecBCD is an enzyme of the E. coli bacterium that initiates recombinational repair from potentially lethal double strand breaks in DNA which may result from ionizing radiation, replication errors, endonucleases, oxidative damage, and a host of other factors. The RecBCD enzyme is both a helicase that unwinds, or separates the strands of DNA, and a nuclease that makes single-stranded nicks in DNA.

RuvABC is a complex of three proteins that mediate branch migration and resolve the Holliday junction created during homologous recombination in bacteria. As such, RuvABC is critical to bacterial DNA repair.
RecQ helicase is a family of helicase enzymes initially found in Escherichia coli that has been shown to be important in genome maintenance. They function through catalyzing the reaction ATP + H2O → ADP + P and thus driving the unwinding of paired DNA and translocating in the 3' to 5' direction. These enzymes can also drive the reaction NTP + H2O → NDP + P to drive the unwinding of either DNA or RNA.

Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolyzing reaction that breaks phosphodiester bonds at either the 3' or the 5' end occurs. Its close relative is the endonuclease, which cleaves phosphodiester bonds in the middle (endo) of a polynucleotide chain. Eukaryotes and prokaryotes have three types of exonucleases involved in the normal turnover of mRNA: 5' to 3' exonuclease (Xrn1), which is a dependent decapping protein; 3' to 5' exonuclease, an independent protein; and poly(A)-specific 3' to 5' exonuclease.

RecA is a 38 kilodalton protein essential for the repair and maintenance of DNA. A RecA structural and functional homolog has been found in every species in which one has been seriously sought and serves as an archetype for this class of homologous DNA repair proteins. The homologous protein is called RAD51 in eukaryotes and RadA in archaea.

Homologous recombination is a type of genetic recombination in which nucleotide sequences are exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids. It is most widely used by cells to accurately repair harmful breaks that occur on both strands of DNA, known as double-strand breaks (DSB). Homologous recombination also produces new combinations of DNA sequences during meiosis, the process by which eukaryotes make gamete cells, like sperm and egg cells in animals. These new combinations of DNA represent genetic variation in offspring, which in turn enables populations to adapt during the course of evolution. Homologous recombination is also used in horizontal gene transfer to exchange genetic material between different strains and species of bacteria and viruses.

The replisome is a complex molecular machine that carries out replication of DNA. The replisome first unwinds double stranded DNA into two single strands. For each of the resulting single strands, a new complementary sequence of DNA is synthesized. The net result is formation of two new double stranded DNA sequences that are exact copies of the original double stranded DNA sequence.
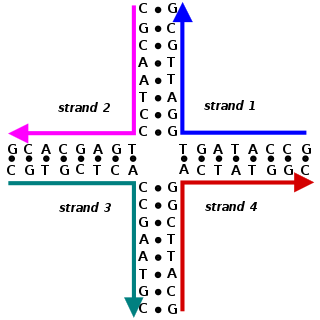
A Holliday junction is a branched nucleic acid structure that contains four double-stranded arms joined together. These arms may adopt one of several conformations depending on buffer salt concentrations and the sequence of nucleobases closest to the junction. The structure is named after Robin Holliday, the molecular biologist who proposed its existence in 1964.

Flap endonuclease 1 is an enzyme that in humans is encoded by the FEN1 gene.
A Chi site or Chi sequence is a short stretch of DNA in the genome of a bacterium near which homologous recombination is more likely to occur than on average across the genome. Chi sites serve as stimulators of DNA double-strand break repair in bacteria, which can arise from radiation or chemical treatments, or result from replication fork breakage during DNA replication. The sequence of the Chi site is unique to each group of closely related organisms; in E. coli and other enteric bacteria, such as Salmonella, the core sequence is 5'-GCTGGTGG-3' plus important nucleotides about 4 to 7 nucleotides to the 3' side of the core sequence. The existence of Chi sites was originally discovered in the genome of bacteriophage lambda, a virus that infects E. coli, but is now known to occur about 1000 times in the E. coli genome.

In genetics, the initial processes involved in repair of a double-strand break by synthesis-dependent strand annealing (SDSA) are identical to those in the double Holliday junction model, and have been most extensively studied in yeast species Saccharomyces cerevisiae. Following a double-stranded break, a protein complex (MRX) binds to either end of the break, working with DNA nucleases to carry out resection, resulting in 5' end digest to produce 3' overhangs of single-stranded DNA. These overhangs are then bound to form a nucleoprotein filament, which can then locate DNA sequences similar to one of the 3' overhangs, initiating a single-stranded strand invasion into the DNA duplex containing these sequences. Once strand invasion has occurred, a displacement loop, or D-loop, is formed, at which point either SDSA or a double Holliday junction occurs.
Stephen Charles Kowalczykowski is a Distinguished Professor of Microbiology and Molecular Genetics at the University of California at Davis. His research focuses on the biochemistry and molecular biology of DNA repair and homologous recombination. His lab combines fluorescence microscopy, optical trapping and microfluidics to manipulate and visualize single molecules of DNA and the enzymes involved in processing and repairing DNA. He calls this scientific approach, "visual biochemistry". Stephen Kowalczykowski was elected to the American Society for Arts and Science in 2005, the National Academy of Sciences in 2007 and was a Harvey Society Lecturer at Rockefeller University in 2012.
Crossover junction endodeoxyribonuclease, also known as Holliday junction resolvase, Holliday junction endonuclease, Holliday junction-cleaving endonuclease, Holliday junction-resolving endoribonuclease, crossover junction endoribonuclease, and cruciform-cutting endonuclease, is an enzyme involved in DNA repair and homologous recombination. Specifically, it performs endonucleolytic cleavage that results in single-stranded crossover between two homologous DNA molecules at the Holliday junction to produce recombinant DNA products for chromosomal segregation. This process is known as Holliday junction resolution.
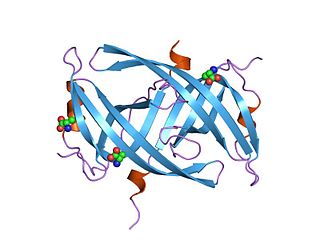
Single-stranded binding proteins (SSBs) are a class of proteins that have been identified in both viruses and organisms from bacteria to humans.
Single-strand DNA-binding protein (SSB) is a protein found in Escherichia coli bacteria, that binds to single-stranded regions of deoxyribonucleic acid (DNA). Single-stranded DNA is produced during all aspects of DNA metabolism: replication, recombination, and repair. As well as stabilizing this single-stranded DNA, SSB proteins bind to and modulate the function of numerous proteins involved in all of these processes.
References
- ↑ Morimatsu, K; Kowalczykowski, SC (22 May 2003). "RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair". Molecular Cell. 11 (5): 1337–1347. doi:10.1016/S1097-2765(03)00188-6. PMID 12769856.
- ↑ Hiom, K (July 2009). "DNA Repair: Common Approaches to Fixing Double-Strand Breaks". Current Biology. 19 (13): R523–R525. doi:10.1016/j.cub.2009.06.009. PMID 19602417.
- 1 2 Handa, N; Morimatsu, K; Lovett, ST; Kowalczykowski, SC (15 May 2009). "Reconstitution of initial steps of dsDNA break repair by the RecF pathway of E. coli". Genes & Development. 23 (10): 1234–1245. doi:10.1101/gad.1780709. PMC 2685532 . PMID 19451222.
- ↑ Kowalczykowski, SC; Clow, J; Somani, R; Varghese, A (5 January 1987). "Effects of the Escherichia coli SSB protein on the binding of Escherichia coli RecA protein to single-stranded DNA. Demonstration of competitive binding and the lack of a specific protein-protein interaction". Journal of Biological Chemistry. 193 (1): 81–95. doi:10.1016/0022-2836(87)90629-2. PMID 3295259.
- 1 2 3 Sakai, A; Cox, MM (30 January 2009). "RecFOR and RecOR as distinct RecA loading pathways" (PDF). Journal of Biological Chemistry. 284 (5): 3264–3272. doi:10.1074/jbc.M807220200. PMC 2631980 . PMID 18986990. Archived from the original (PDF) on 17 June 2010.
- ↑ Inoue, H; Honda, M; Ikawa, S; Shibata, T; Mikawa, T (January 2008). "The process of displacing the single-stranded DNA-binding protein from single-stranded DNA by RecO and RecR proteins". Nucleic Acids Research. 36 (1): 94–109. doi:10.1093/nar/gkm1004. PMC 2248737 . PMID 18000001.