
Chromosomal crossover, or crossing over, is the exchange of genetic material during sexual reproduction between two homologous chromosomes' non-sister chromatids that results in recombinant chromosomes. It is one of the final phases of genetic recombination, which occurs in the pachytene stage of prophase I of meiosis during a process called synapsis. Synapsis begins before the synaptonemal complex develops and is not completed until near the end of prophase I. Crossover usually occurs when matching regions on matching chromosomes break and then reconnect to the other chromosome.

Helicases are a class of enzymes thought to be vital to all organisms. Their main function is to unpack an organism's genetic material. Helicases are motor proteins that move directionally along a nucleic acid phosphodiester backbone, separating two hybridized nucleic acid strands, using energy from ATP hydrolysis. There are many helicases, representing the great variety of processes in which strand separation must be catalyzed. Approximately 1% of eukaryotic genes code for helicases.

RecBCD is an enzyme of the E. coli bacterium that initiates recombinational repair from potentially lethal double strand breaks in DNA which may result from ionizing radiation, replication errors, endonucleases, oxidative damage, and a host of other factors. The RecBCD enzyme is both a helicase that unwinds, or separates the strands of DNA, and a nuclease that makes single-stranded nicks in DNA.

RuvABC is a complex of three proteins that mediate branch migration and resolve the Holliday junction created during homologous recombination in bacteria. As such, RuvABC is critical to bacterial DNA repair.
RecQ helicase is a family of helicase enzymes initially found in Escherichia coli that has been shown to be important in genome maintenance. They function through catalyzing the reaction ATP + H2O → ADP + P and thus driving the unwinding of paired DNA and translocating in the 3' to 5' direction. These enzymes can also drive the reaction NTP + H2O → NDP + P to drive the unwinding of either DNA or RNA.

Sulfolobus is a genus of microorganism in the family Sulfolobaceae. It belongs to the archaea domain.

Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids. It is widely used by cells to accurately repair harmful breaks that occur on both strands of DNA, known as double-strand breaks (DSB), in a process called homologous recombinational repair (HRR). Homologous recombination also produces new combinations of DNA sequences during meiosis, the process by which eukaryotes make gamete cells, like sperm and egg cells in animals. These new combinations of DNA represent genetic variation in offspring, which in turn enables populations to adapt during the course of evolution. Homologous recombination is also used in horizontal gene transfer to exchange genetic material between different strains and species of bacteria and viruses.

Werner syndrome ATP-dependent helicase, also known as DNA helicase, RecQ-like type 3, is an enzyme that in humans is encoded by the WRN gene. WRN is a member of the RecQ Helicase family. Helicase enzymes generally unwind and separate double-stranded DNA. These activities are necessary before DNA can be copied in preparation for cell division. Helicase enzymes are also critical for making a blueprint of a gene for protein production, a process called transcription. Further evidence suggests that Werner protein plays a critical role in repairing DNA. Overall, this protein helps maintain the structure and integrity of a person's DNA.
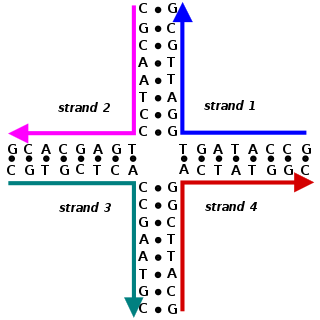
A Holliday junction is a branched nucleic acid structure that contains four double-stranded arms joined. These arms may adopt one of several conformations depending on buffer salt concentrations and the sequence of nucleobases closest to the junction. The structure is named after Robin Holliday, the molecular biologist who proposed its existence in 1964.

In taxonomy, the Sulfolobales are an order of the Thermoprotei.

Bloom syndrome protein is a protein that in humans is encoded by the BLM gene and is not expressed in Bloom syndrome.

Crossover junction endonuclease MUS81 is an enzyme that in humans is encoded by the MUS81 gene.
Fanconi anemia, complementation group M, also known as FANCM is a human gene. It is an emerging target in cancer therapy, in particular cancers with specific genetic deficiencies.
A Chi site or Chi sequence is a short stretch of DNA in the genome of a bacterium near which homologous recombination is more likely to occur than on average across the genome. Chi sites serve as stimulators of DNA double-strand break repair in bacteria, which can arise from radiation or chemical treatments, or result from replication fork breakage during DNA replication. The sequence of the Chi site is unique to each group of closely related organisms; in E. coli and other enteric bacteria, such as Salmonella, the core sequence is 5'-GCTGGTGG-3' plus important nucleotides about 4 to 7 nucleotides to the 3' side of the core sequence. The existence of Chi sites was originally discovered in the genome of bacteriophage lambda, a virus that infects E. coli, but is now known to occur about 1000 times in the E. coli genome.
The RecF pathway, also called the RecFOR pathway, is a pathway of homologous recombination that repairs DNA in bacteria. It repairs breaks that occur on only one of DNA's two strands, known as single-strand gaps. The RecF pathway can also repair double-strand breaks in DNA when the RecBCD pathway, another pathway of homologous recombination in bacteria, is inactivated by mutations. Like the RecBCD pathway, the RecF pathway requires RecA for strand invasion. The two pathways are also similar in their phases of branch migration, in which the Holliday junction slides in one direction, and resolution, in which the Holliday junctions are cleaved apart by enzymes.

Synthesis-dependent strand annealing (SDSA) is a major mechanism of homology-directed repair of DNA double-strand breaks (DSBs). Although many of the features of SDSA were first suggested in 1976, the double-Holliday junction model proposed in 1983 was favored by many researchers. In 1994, studies of double-strand gap repair in Drosophila were found to be incompatible with the double-Holliday junction model, leading researchers to propose a model they called synthesis-dependent strand annealing. Subsequent studies of meiotic recombination in S. cerevisiae found that non-crossover products appear earlier than double-Holliday junctions or crossover products, challenging the previous notion that both crossover and non-crossover products are produced by double-Holliday junctions and leading the authors to propose that non-crossover products are generated through SDSA.
Crossover junction endodeoxyribonuclease, also known as Holliday junction resolvase, Holliday junction endonuclease, Holliday junction-cleaving endonuclease, Holliday junction-resolving endoribonuclease, crossover junction endoribonuclease, and cruciform-cutting endonuclease, is an enzyme involved in DNA repair and homologous recombination. Specifically, it performs endonucleolytic cleavage that results in single-stranded crossover between two homologous DNA molecules at the Holliday junction to produce recombinant DNA products for chromosomal segregation. This process is known as Holliday junction resolution.
Sulfolobus acidocaldarius is a thermoacidophilic archaeon that belongs to the phylum Thermoproteota. S. acidocaldarius was the first Sulfolobus species to be described, in 1972 by Thomas D. Brock and collaborators. This species was found to grow optimally between 75 and 80 °C, with pH optimum in the range of 2-3.

Helicase, POLQ-like, also known as helicase Q, hel308 and Holliday junction migration protein, encoded by the gene HELQ1, is a DNA helicase found in humans, archea and many other organisms.
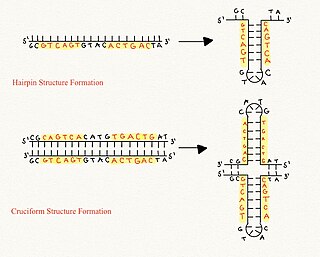
Cruciform DNA is a form of non-B DNA, or an alternative DNA structure. The formation of cruciform DNA requires the presence of palindromes called inverted repeat sequences. These inverted repeats contain a sequence of DNA in one strand that is repeated in the opposite direction on the other strand. As a result, inverted repeats are self-complementary and can give rise to structures such as hairpins and cruciforms. Cruciform DNA structures require at least a six nucleotide sequence of inverted repeats to form a structure consisting of a stem, branch point and loop in the shape of a cruciform, stabilized by negative DNA supercoiling.















