Related Research Articles

Phosphoglucomutase is an enzyme that transfers a phosphate group on an α-D-glucose monomer from the 1 to the 6 position in the forward direction or the 6 to the 1 position in the reverse direction.
Phosphofructokinase-2 (6-phosphofructo-2-kinase, PFK-2) or fructose bisphosphatase-2 (FBPase-2), is an enzyme indirectly responsible for regulating the rates of glycolysis and gluconeogenesis in cells. It catalyzes formation and degradation of a significant allosteric regulator, fructose-2,6-bisphosphate (Fru-2,6-P2) from substrate fructose-6-phosphate. Fru-2,6-P2 contributes to the rate-determining step of glycolysis as it activates enzyme phosphofructokinase 1 in the glycolysis pathway, and inhibits fructose-1,6-bisphosphatase 1 in gluconeogenesis. Since Fru-2,6-P2 differentially regulates glycolysis and gluconeogenesis, it can act as a key signal to switch between the opposing pathways. Because PFK-2 produces Fru-2,6-P2 in response to hormonal signaling, metabolism can be more sensitively and efficiently controlled to align with the organism's glycolytic needs. This enzyme participates in fructose and mannose metabolism. The enzyme is important in the regulation of hepatic carbohydrate metabolism and is found in greatest quantities in the liver, kidney and heart. In mammals, several genes often encode different isoforms, each of which differs in its tissue distribution and enzymatic activity. The family described here bears a resemblance to the ATP-driven phospho-fructokinases; however, they share little sequence similarity, although a few residues seem key to their interaction with fructose 6-phosphate.

Prostatic acid phosphatase (PAP), also prostatic specific acid phosphatase (PSAP), is an enzyme produced by the prostate. It may be found in increased amounts in men who have prostate cancer or other diseases.

A phytase is any type of phosphatase enzyme that catalyzes the hydrolysis of phytic acid – an indigestible, organic form of phosphorus that is found in many plant tissues, especially in grains and oil seeds – and releases a usable form of inorganic phosphorus. While phytases have been found to occur in animals, plants, fungi and bacteria, phytases have been most commonly detected and characterized from fungi.

Thromboxane A synthase 1 , also known as TBXAS1, is a cytochrome P450 enzyme that, in humans, is encoded by the TBXAS1 gene.

4-Hydroxyphenylpyruvate dioxygenase (HPPD), also known as α-ketoisocaproate dioxygenase, is an Fe(II)-containing non-heme oxygenase that catalyzes the second reaction in the catabolism of tyrosine - the conversion of 4-hydroxyphenylpyruvate into homogentisate. HPPD also catalyzes the conversion of phenylpyruvate to 2-hydroxyphenylacetate and the conversion of α-ketoisocaproate to β-hydroxy β-methylbutyrate. HPPD is an enzyme that is found in nearly all aerobic forms of life.
Acid phosphatase is an enzyme that frees attached phosphoryl groups from other molecules during digestion. It can be further classified as a phosphomonoesterase. It is stored in lysosomes and functions when these fuse with endosomes, which are acidified while they function; therefore, it has an acid pH optimum. This enzyme is present in many animal and plant species.
Purine nucleoside phosphorylase, PNP, PNPase or inosine phosphorylase is an enzyme that in humans is encoded by the NP gene. It catalyzes the chemical reaction
Phospholipase D (EC 3.1.4.4, lipophosphodiesterase II, lecithinase D, choline phosphatase, PLD; systematic name phosphatidylcholine phosphatidohydrolase) is an enzyme of the phospholipase superfamily that catalyses the following reaction

Beta-hexosaminidase subunit beta is an enzyme that in humans is encoded by the HEXB gene.
Tartrate-resistant acid phosphatase, also called acid phosphatase 5, tartrate resistant (ACP5), is a glycosylated monomeric metalloprotein enzyme expressed in mammals. It has a molecular weight of approximately 35kDa, a basic isoelectric point (7.6–9.5), and optimal activity in acidic conditions. TRAP is synthesized as latent proenzyme and activated by proteolytic cleavage and reduction. It is differentiated from other mammalian acid phosphatases by its resistance to inhibition by tartrate and by its molecular weight.
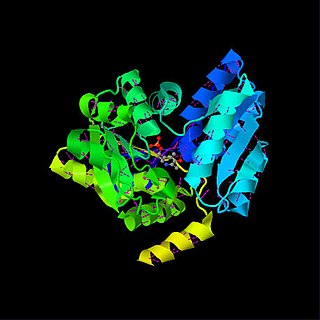
Serine dehydratase or L-serine ammonia lyase (SDH) is in the β-family of pyridoxal phosphate-dependent (PLP) enzymes. SDH is found widely in nature, but its structure and properties vary among species. SDH is found in yeast, bacteria, and the cytoplasm of mammalian hepatocytes. SDH catalyzes the deamination of L-serine to yield pyruvate, with the release of ammonia.
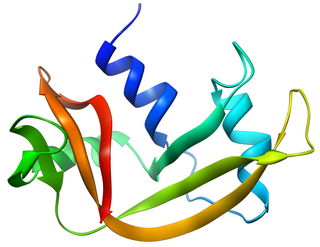
Pancreatic ribonuclease family is a superfamily of pyrimidine-specific endonucleases found in high quantity in the pancreas of certain mammals and of some reptiles.
Deoxyribonuclease IV (phage-T4-induced) is catalyzes the degradation nucleotides in DsDNA by attacking the 5'-terminal end.

Malate dehydrogenase (oxaloacetate-decarboxylating) (NADP+) (EC 1.1.1.40) or NADP-malic enzyme (NADP-ME) is an enzyme that catalyzes the chemical reaction in the presence of a bivalent metal ion:

Serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform is an enzyme that is encoded by the PPP2CA gene.

Low molecular weight phosphotyrosine protein phosphatase is an enzyme that in humans is encoded by the ACP1 gene.

The enzyme phosphatidate phosphatase (PAP, EC 3.1.3.4) is a key regulatory enzyme in lipid metabolism, catalyzing the conversion of phosphatidate to diacylglycerol:
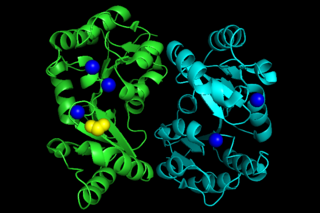
Phosphoglycolate phosphatase(EC 3.1.3.18; systematic name 2-phosphoglycolate phosphohydrolase), also commonly referred to as phosphoglycolate hydrolase, 2-phosphoglycolate phosphatase, P-glycolate phosphatase, and phosphoglycollate phosphatase, is an enzyme responsible for catalyzing the conversion of 2-phosphoglycolate into glycolate and phosphate:

In enzymology, a polynucleotide adenylyltransferase is an enzyme that catalyzes the chemical reaction
References
- ↑ B. C. Antanaitis; P. Aisen (1983). "Uteroferrin and the purple acid phosphatases". Advances in Inorganic Biochemistry . 5: 111–136. PMID 6382957.
- ↑ David C. Schlosnagle; Fuller W. Bazer; John C. M. Tsibris; R. Michael Roberts (December 1974). "An iron-containing phosphatase induced by progesterone in the uterine fluids of pigs". Journal of Biological Chemistry . 249 (23): 7574–9. doi: 10.1016/S0021-9258(19)81276-0 . PMID 4373472.
- ↑ Antanaitis BC, Aisen P (February 1984). "Stoichiometry of iron binding by uteroferrin and its relationship to phosphate content". Journal of Biological Chemistry . 259 (4): 2066–2069. doi: 10.1016/S0021-9258(17)43315-1 . PMID 6698956.
- ↑ Bruce P. Gaber; James P. Sheridan; Fuller W. Bazer; R. Michael Roberts (September 1979). "Resonance Raman scattering from uteroferrin, the purple glycoprotein of the porcine uterus". Journal of Biological Chemistry . 254 (17): 8340–8342. doi: 10.1016/S0021-9258(19)86895-3 . PMID 468828.
- ↑ Jussi M. Halleen; Helena Kaija; Jan J. Stepan; Pirkko Vihko; H. Kalervo Väänänen (April 1998). "Studies on the protein tyrosine phosphatase activity of tartrate-resistant acid phosphatase". Archives of Biochemistry and Biophysics . 352 (1): 97–102. doi:10.1006/abbi.1998.0600. PMID 9521821.
- ↑ Deirdre K. Lord; Nicholas C. P. Cross; Maria A. Bevilacqua; Susan H. Rider; Patricia A. Gorman; Ann V. Groves; Donald W. Moss; Denise Sheer; Timothy M. Cox (April 1990). "Type 5 acid phosphatase. Sequence, expression and chromosomal localization of a differentiation-associated protein of the human macrophage". European Journal of Biochemistry . 189 (2): 287–293. doi: 10.1111/j.1432-1033.1990.tb15488.x . PMID 2338077.
- ↑ Gerhard Schenk; Yubin Ge; Lyle E. Carrington; Ceridwen J. Wynne; Iain R. Searle; Bernard J. Carroll; Susan Hamilton; John de Jersey (October 1999). "Binuclear metal centers in plant purple acid phosphatases: Fe-Mn in sweet potato and Fe-Zn in soybean" (PDF). Archives of Biochemistry and Biophysics . 370 (2): 183–189. doi:10.1006/abbi.1999.1407. PMID 10510276.
- ↑ Thomas Klabunde; Norbert Sträter; Bernt Krebs; Herbert Witzel (June 1995). "Structural relationship between the mammalian Fe(III)-Fe(II) and the Fe(III)-Zn(II) plant purple acid phosphatases". FEBS Letters . 367 (1): 56–60. doi: 10.1016/0014-5793(95)00536-I . PMID 7601285.
- ↑ Gerhard Schenk; Michael L. J. Korsinczky; David A. Hume; Susan Hamilton; John DeJersey (September 2000). "Purple acid phosphatases from bacteria: similarities to mammalian and plant enzymes" (PDF). Gene . 255 (2): 419–424. doi:10.1016/S0378-1119(00)00305-X. PMID 11024303.
- ↑ Barbro Ek-Rylander; Per Bill; Maria Norgård; Stefan Nilsson; Göran Andersson (December 1991). "Cloning, sequence, and developmental expression of a type 5, tartrate-resistant, acid phosphatase of rat bone". Journal of Biological Chemistry . 266 (36): 24684–24689. doi: 10.1016/S0021-9258(18)54284-8 . PMID 1722212.
- ↑ Ping Ling; R. Michael Roberts (April 1993). "Uteroferrin and intracellular tartrate-resistant acid phosphatases are the products of the same gene". Journal of Biological Chemistry . 268 (10): 6896–6902. doi: 10.1016/S0021-9258(18)53124-0 . PMID 8463220.