Related Research Articles

Enterobacteria phage λ is a bacterial virus, or bacteriophage, that infects the bacterial species Escherichia coli. It was discovered by Esther Lederberg in 1950. The wild type of this virus has a temperate life cycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase, during which it kills and lyses the cell to produce offspring. Lambda strains, mutated at specific sites, are unable to lysogenize cells; instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell.
A regulatory sequence is a segment of a nucleic acid molecule which is capable of increasing or decreasing the expression of specific genes within an organism. Regulation of gene expression is an essential feature of all living organisms and viruses.

DnaA is a protein that activates initiation of DNA replication in bacteria. Based on the Replicon Model, a positively active initiator molecule contacts with a particular spot on a circular chromosome called the replicator to start DNA replication. It is a replication initiation factor which promotes the unwinding of DNA at oriC. The DnaA proteins found in all bacteria engage with the DnaA boxes to start chromosomal replication. The onset of the initiation phase of DNA replication is determined by the concentration of DnaA. DnaA accumulates during growth and then triggers the initiation of replication. Replication begins with active DnaA binding to 9-mer (9-bp) repeats upstream of oriC. Binding of DnaA leads to strand separation at the 13-mer repeats. This binding causes the DNA to loop in preparation for melting open by the helicase DnaB.

The SOS response is a global response to DNA damage in which the cell cycle is arrested and DNA repair and mutagenesis are induced. The system involves the RecA protein. The RecA protein, stimulated by single-stranded DNA, is involved in the inactivation of the repressor (LexA) of SOS response genes thereby inducing the response. It is an error-prone repair system that contributes significantly to DNA changes observed in a wide range of species.
Integrons are genetic mechanisms that allow bacteria to adapt and evolve rapidly through the stockpiling and expression of new genes. These genes are embedded in a specific genetic structure called gene cassette that generally carries one promoterless open reading frame (ORF) together with a recombination site (attC). Integron cassettes are incorporated to the attI site of the integron platform by site-specific recombination reactions mediated by the integrase.

RecA is a 38 kilodalton protein essential for the repair and maintenance of DNA in bacteria. Structural and functional homologs to RecA have been found in all kingdoms of life. RecA serves as an archetype for this class of homologous DNA repair proteins. The homologous protein is called RAD51 in eukaryotes and RadA in archaea.
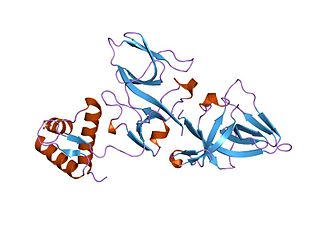
The LexA repressor or LexA is a transcriptional repressor that represses SOS response genes coding primarily for error-prone DNA polymerases, DNA repair enzymes and cell division inhibitors. LexA forms de facto a two-component regulatory system with RecA, which senses DNA damage at stalled replication forks, forming monofilaments and acquiring an active conformation capable of binding to LexA and causing LexA to cleave itself, in a process called autoproteolysis.

General transcription factors (GTFs), also known as basal transcriptional factors, are a class of protein transcription factors that bind to specific sites (promoter) on DNA to activate transcription of genetic information from DNA to messenger RNA. GTFs, RNA polymerase, and the mediator constitute the basic transcriptional apparatus that first bind to the promoter, then start transcription. GTFs are also intimately involved in the process of gene regulation, and most are required for life.
Cis-regulatory elements (CREs) or cis-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphogenesis, the development of anatomy, and other aspects of embryonic development, studied in evolutionary developmental biology.
Gene structure is the organisation of specialised sequence elements within a gene. Genes contain most of the information necessary for living cells to survive and reproduce. In most organisms, genes are made of DNA, where the particular DNA sequence determines the function of the gene. A gene is transcribed (copied) from DNA into RNA, which can either be non-coding (ncRNA) with a direct function, or an intermediate messenger (mRNA) that is then translated into protein. Each of these steps is controlled by specific sequence elements, or regions, within the gene. Every gene, therefore, requires multiple sequence elements to be functional. This includes the sequence that actually encodes the functional protein or ncRNA, as well as multiple regulatory sequence regions. These regions may be as short as a few base pairs, up to many thousands of base pairs long.
In molecular genetics, a regulon is a group of genes that are regulated as a unit, generally controlled by the same regulatory gene that expresses a protein acting as a repressor or activator. This terminology is generally, although not exclusively, used in reference to prokaryotes, whose genomes are often organized into operons; the genes contained within a regulon are usually organized into more than one operon at disparate locations on the chromosome. Applied to eukaryotes, the term refers to any group of non-contiguous genes controlled by the same regulatory gene.
In genetics and cell biology, repression is a mechanism often used to decrease or inhibit the expression of a gene. Removal of repression is called derepression. This mechanism may occur at different stages in the expression of a gene, with the result of increasing the overall RNA or protein products. Dysregulation of derepression mechanisms can result in altered gene expression patterns, which may lead to negative phenotypic consequences such as disease.
The gal operon is a prokaryotic operon, which encodes enzymes necessary for galactose metabolism. Repression of gene expression for this operon works via binding of repressor molecules to two operators. These repressors dimerize, creating a loop in the DNA. The loop as well as hindrance from the external operator prevent RNA polymerase from binding to the promoter, and thus prevent transcription. Additionally, since the metabolism of galactose in the cell is involved in both anabolic and catabolic pathways, a novel regulatory system using two promoters for differential repression has been identified and characterized within the context of the gal operon.
The SymE-SymR toxin-antitoxin system consists of a small symbiotic endonuclease toxin, SymE, and a non-coding RNA symbiotic RNA antitoxin, SymR, which inhibits SymE translation. SymE-SymR is a type I toxin-antitoxin system, and is under regulation by the antitoxin, SymR. The SymE-SymR complex is believed to play an important role in recycling damaged RNA and DNA. The relationship and corresponding structures of SymE and SymR provide insight into the mechanism of toxicity and overall role in prokaryotic systems.
DNA Polymerase V is a polymerase enzyme involved in DNA repair mechanisms in bacteria, such as Escherichia coli. It is composed of a UmuD' homodimer and a UmuC monomer, forming the UmuD'2C protein complex. It is part of the Y-family of DNA Polymerases, which are capable of performing DNA translesion synthesis (TLS). Translesion polymerases bypass DNA damage lesions during DNA replication - if a lesion is not repaired or bypassed the replication fork can stall and lead to cell death. However, Y polymerases have low sequence fidelity during replication. When the UmuC and UmuD' proteins were initially discovered in E. coli, they were thought to be agents that inhibit faithful DNA replication and caused DNA synthesis to have high mutation rates after exposure to UV-light. The polymerase function of Pol V was not discovered until the late 1990s when UmuC was successfully extracted, consequent experiments unequivocally proved UmuD'2C is a polymerase. This finding lead to the detection of many Pol V orthologs and the discovery of the Y-family of polymerases.
The nik operon is an operon required for uptake of nickel ions into the cell. It is present in many bacteria, but has been extensively studied in Helicobacter pylori. Nickel is an essential nutrient for many microorganisms, where it participates in a variety of cellular processes. However, excessive levels of nickel ions in cell can be fatal to the cell. Nickel ion concentration in the cell is regulated through the nik operon.
The gene rpoN encodes the sigma factor sigma-54, a protein in Escherichia coli and other species of bacteria. RpoN antagonizes RpoS sigma factors.
Q-system is a genetic tool that allows to express transgenes in a living organism. Originally the Q-system was developed for use in the vinegar fly Drosophila melanogaster, and was rapidly adapted for use in cultured mammalian cells, zebrafish, worms and mosquitoes. The Q-system utilizes genes from the qa cluster of the bread fungus Neurospora crassa, and consists of four components: the transcriptional activator (QF/QF2/QF2w), the enhancer QUAS, the repressor QS, and the chemical de-repressor quinic acid. Similarly to GAL4/UAS and LexA/LexAop, the Q-system is a binary expression system that allows to express reporters or effectors in a defined subpopulation of cells with the purpose of visualising these cells or altering their function. In addition, GAL4/UAS, LexA/LexAop and the Q-system function independently of each other and can be used simultaneously to achieve a desired pattern of reporter expression, or to express several reporters in different subsets of cells.
The locus of enterocyte effacement-encoded regulator (Ler) is a regulatory protein that controls bacterial pathogenicity of enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic Escherichia coli (EHEC). More specifically, Ler regulates the locus of enterocyte effacement (LEE) pathogenicity island genes, which are responsible for creating intestinal attachment and effacing lesions and subsequent diarrhea: LEE1, LEE2, and LEE3. LEE1, 2, and 3 carry the information necessary for a type III secretion system. The transcript encoding the Ler protein is the open reading frame 1 on the LEE1 operon.

Ivan Erill is a Spanish computational biologist known for his research in comparative genomics and molecular microbiology. His work focuses primarily on bacterial comparative genomics, through the development of computational methods for analyzing regulatory networks and their evolution.
References
- ↑ Henkin, Tina M.; Peters, Joseph E. (2020). Snyder and Champness molecular genetics of bacteria (Fifth ed.). Hoboken, NJ : Washington, D.C: John Wiley & Sons, Inc. p. 409. ISBN 9781555819750.
- ↑ Walker, Graham C. (October 1995). "SOS-regulated proteins in translesion DNA synthesis and mutagenesis". Trends in Biochemical Sciences. 20 (10): 416–20. doi:10.1016/s0968-0004(00)89091-x. PMID 8533155.
- ↑ Gillor, Osnat; Vriezen, Jan A. C.; Riley, Margaret A. (Jun 2008). "The role of SOS boxes in enteric bacteriocin regulation". Microbiology. 154 (6): 1783–1792. doi: 10.1099/mic.0.2007/016139-0 . PMC 2729051 . PMID 18524933.
- ↑ Fernández de Henestrosa, Antonio R.; Rivera, Eusebi; Tapias, Angels; Barbé, Jordi (June 1998). "Identification of the Rhodobacter sphaeroides SOS box". Molecular Microbiology. 28 (5): 991–1003. doi: 10.1046/j.1365-2958.1998.00860.x . PMID 9663685.
- ↑ Tapias, A.; Barbé, J. (August 1999). "Regulation of divergent transcription from the uvrA-ssb promoters in Sinorhizobium meliloti". Molecular and General Genetics MGG. 262 (1): 121–130. doi:10.1007/s004380051066. PMID 10503543. S2CID 2373618.
- ↑ Erill, Ivan; Escribano, Marcos; Campoy, Susana; Barbé, Jordi (22 November 2003). "In silico analysis reveals substantial variability in the gene contents of the gamma proteobacteria LexA-regulon". Bioinformatics. 19 (17): 2225–2236. doi: 10.1093/bioinformatics/btg303 . PMID 14630651.
- ↑ Campoy, Susana; Fontes, Marta; Padmanabhan, S.; Cortés, Pilar; Llagostera, Montserrat; Barbé, Jordi (August 2003). "LexA-independent DNA damage-mediated induction of gene expression in Myxococcus xanthus". Molecular Microbiology. 49 (3): 769–781. doi:10.1046/j.1365-2958.2003.03592.x. PMID 12864858.
- ↑ Winterling, Kevin W.; Chafin, David; Hayes, Jeffery J.; Sun, Ji; Levine, Arthur S.; Yasbin, Ronald E.; Woodgate, Roger (15 April 1998). "The Bacillus subtilis DinR Binding Site: Redefinition of the Consensus Sequence". Journal of Bacteriology. 180 (8): 2201–2211. doi:10.1128/JB.180.8.2201-2211.1998. PMC 107149 . PMID 9555905.
- ↑ Davis, Elaine O.; Dullaghan, Edith M.; Rand, Lucinda (15 June 2002). "Definition of the Mycobacterial SOS Box and Use To Identify LexA-Regulated Genes in Mycobacterium tuberculosis". Journal of Bacteriology. 184 (12): 3287–3295. doi:10.1128/JB.184.12.3287-3295.2002. PMC 135081 . PMID 12029045.
- ↑ Mazón, G.; Lucena, J. M.; Campoy, S.; Fernández de Henestrosa, A. R.; Candau, P.; Barbé, J. (February 2004). "LexA-binding sequences in Gram-positive and cyanobacteria are closely related". Molecular Genetics and Genomics. 271 (1): 40–49. doi:10.1007/s00438-003-0952-x. PMID 14652736. S2CID 9219764.