
Pseudomonadota is a major phylum of Gram-negative bacteria. Currently, they are considered the predominant phylum within the realm of bacteria. They are naturally found as pathogenic and free-living (non-parasitic) genera. The phylum comprises six classes Acidithiobacillia, Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, Hydrogenophilia, and Zetaproteobacteria. The Pseudomonadota are widely diverse, with differences in morphology, metabolic processes, relevance to humans, and ecological influence.
The Rhodospirillales are an order of Pseudomonadota.
The Chloroflexia are a class of bacteria in the phylum Chloroflexota. Chloroflexia are typically filamentous, and can move about through bacterial gliding. It is named after the order Chloroflexales.

The Chlamydiota are a bacterial phylum and class whose members are remarkably diverse, including pathogens of humans and animals, symbionts of ubiquitous protozoa, and marine sediment forms not yet well understood. All of the Chlamydiota that humans have known about for many decades are obligate intracellular bacteria; in 2020 many additional Chlamydiota were discovered in ocean-floor environments, and it is not yet known whether they all have hosts. Historically it was believed that all Chlamydiota had a peptidoglycan-free cell wall, but studies in the 2010s demonstrated a detectable presence of peptidoglycan, as well as other important proteins.

"Candidatus Pelagibacter", with the single species "Ca. P. communis", was isolated in 2002 and given a specific name, although it has not yet been described as required by the bacteriological code. It is an abundant member of the SAR11 clade in the phylum Alphaproteobacteria. SAR11 members are highly dominant organisms found in both salt and fresh water worldwide and were originally known only from their rRNA genes, first identified in the Sargasso Sea in 1990 by Stephen Giovannoni's laboratory at Oregon State University and later found in oceans worldwide. "Ca. P. communis" and its relatives may be the most abundant organisms in the ocean, and quite possibly the most abundant bacteria in the entire world. It can make up about 25% of all microbial plankton cells, and in the summer they may account for approximately half the cells present in temperate ocean surface water. The total abundance of "Ca. P. communis" and relatives is estimated to be about 2 × 1028 microbes.

The Rickettsiales, informally called rickettsias, are an order of small Alphaproteobacteria. They are obligate intracellular parasites, and some are notable pathogens, including Rickettsia, which causes a variety of diseases in humans, and Ehrlichia, which causes diseases in livestock. Another genus of well-known Rickettsiales is the Wolbachia, which infect about two-thirds of all arthropods and nearly all filarial nematodes. Genetic studies support the endosymbiotic theory according to which mitochondria and related organelles developed from members of this group.

The Hyphomicrobiales are an order of Gram-negative Alphaproteobacteria.

Sphingomonadaceae are a gram-negative bacterial family of the Alphaproteobacteria. An important feature is the presence of sphingolipids in the outer membrane of the cell wall. The cells are ovoid or rod-shaped. Others are also pleomorphic, i.e. the cells change the shape over time. Some species from Sphingomonadaceae family are dominant components of biofilms.

The Rickettsiaceae are a family of bacteria. The genus Rickettsia is the most prominent genus within the family. The bacteria that eventually formed the mitochondrion is believed to have originated from this family. Most human pathogens in this family are in genus Rickettsia. They spend part of their lifecycle in the bodies of arthropods such as ticks or lice, and are then transmitted to humans or other mammals by the bite of the arthropod. It contains Gram-negative bacteria, very sensitive to environmental exposure, thus is adapted to obligate intracellular infection. Rickettsia rickettsii is considered the prototypical infectious organism in the group.
The Holosporaceae are a family of bacteria. The member Holospora is an intracellular parasite found in the unicellular protozoa Paramecium.
The proto-mitochondrion is the hypothetical ancestral bacterial endosymbiont from which all mitochondria in eukaryotes are thought to descend, after an episode of symbiogenesis which created the aerobic eukaryotes.
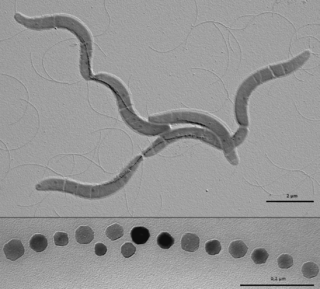
"Candidatus Midichloria" is a candidatus genus of Gram-negative, non-endospore-forming bacteria, with a bacillus shape around 0.45 μm in diameter and 1.2 μm in length. First described in 2004 with the temporary name IricES1, "Candidatus Midichloria" species are symbionts of several species of hard ticks. They live in the cells of the ovary of the females of this tick species. These bacteria have been observed in the mitochondria of the host cells, a trait that has never been described in any other symbiont of animals.

Gracilicutes is a clade in bacterial phylogeny.

The Pelagibacterales are an order in the Alphaproteobacteria composed of free-living marine bacteria that make up roughly one in three cells at the ocean's surface. Overall, members of the Pelagibacterales are estimated to make up between a quarter and a half of all prokaryotic cells in the ocean.
Nitrospirota is a phylum of bacteria. It includes multiple genera, such as Nitrospira, the largest. The first member of this phylum, Nitrospira marina, was discovered in 1985. The second member, Nitrospira moscoviensis, was discovered in 1995.
There are several models of the Branching order of bacterial phyla, one of these was proposed in 1987 paper by Carl Woese.
Nanohaloarchaea is a clade of diminutive archaea with small genomes and limited metabolic capabilities, belonging to the DPANN archaea. They are ubiquitous in hypersaline habitats, which they share with the extremely halophilic haloarchaea.
The Pelagibacteraceae are a family in the Alphaproteobacteria composed of free-living marine bacteria.
The Magnetococcales were an order of Alphaproteobacteria, but now the mitochondria are considered as sister to the alphaproteobactera, together forming the sister the marineproteo1 group, together forming the sister to Magnetococcidae.
Asgard or Asgardarchaeota is a proposed superphylum belonging to the domain Archaea that contain eukaryotic signature proteins. It appears that the eukaryotes, the domain that contains the animals, plants, and fungi, emerged within the Asgard, in a branch containing the Heimdallarchaeota. This supports the two-domain system of classification over the three-domain system.









