| Rickettsiales | |
|---|---|
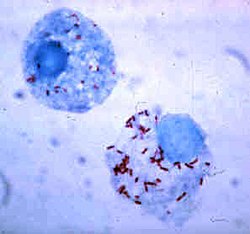 | |
| Rickettsia rickettsii (red dots) in the cell of a deer tick | |
| Scientific classification | |
| Domain: | Bacteria |
| Kingdom: | Pseudomonadati |
| Phylum: | Pseudomonadota |
| Class: | Alphaproteobacteria |
| Subclass: | "Rickettsidae" |
| Order: | Rickettsiales Gieszczykiewicz 1939 (Approved Lists 1980) |
| Families | |
| |
The Rickettsiales, informally called rickettsias, are an order of small Alphaproteobacteria. They are obligate intracellular parasites, and some are notable pathogens, including Rickettsia , which causes a variety of diseases in humans, and Ehrlichia , which causes diseases in livestock. Another genus of well-known Rickettsiales is the Wolbachia , which infect about two-thirds of all arthropods and nearly all filarial nematodes. [2] Genetic studies support the endosymbiotic theory according to which mitochondria and related organelles developed from members of this group. [3] However, more recent examination has suggested that the mitochondria and Rickettsiales diverged independently. [4]
Contents
- Rickettsiales phylogeny
- Phylogenetic relationship between Rickettsiales and Pelagibacterales (SAR11)
- Reductive evolution
- References
The Rickettsiales are difficult to culture, as they rely on living eukaryotic host cells for their survival.