Nitrogen fixation is a chemical process by which molecular nitrogen (N
2), with a strong triple covalent bond, in the air is converted into ammonia (NH
3) or related nitrogenous compounds, typically in soil or aquatic systems but also in industry. Atmospheric nitrogen is molecular dinitrogen, a relatively nonreactive molecule that is metabolically useless to all but a few microorganisms. Biological nitrogen fixation or diazotrophy is an important microbially mediated process that converts dinitrogen (N2) gas to ammonia (NH3) using the nitrogenase protein complex (Nif).

Leghemoglobin is an oxygen-carrying phytoglobin found in the nitrogen-fixing root nodules of leguminous plants. It is produced by these plants in response to the roots being colonized by nitrogen-fixing bacteria, termed rhizobia, as part of the symbiotic interaction between plant and bacterium: roots not colonized by Rhizobium do not synthesise leghemoglobin. Leghemoglobin has close chemical and structural similarities to hemoglobin, and, like hemoglobin, is red in colour. It was originally thought that the heme prosthetic group for plant leghemoglobin was provided by the bacterial symbiont within symbiotic root nodules. However, subsequent work shows that the plant host strongly expresses heme biosynthesis genes within nodules, and that activation of those genes correlates with leghemoglobin gene expression in developing nodules.

Rhizobia are diazotrophic bacteria that fix nitrogen after becoming established inside the root nodules of legumes (Fabaceae). To express genes for nitrogen fixation, rhizobia require a plant host; they cannot independently fix nitrogen. In general, they are gram negative, motile, non-sporulating rods.

Rhizobium is a genus of Gram-negative soil bacteria that fix nitrogen. Rhizobium species form an endosymbiotic nitrogen-fixing association with roots of (primarily) legumes and other flowering plants.

Ensifer meliloti are an aerobic, Gram-negative, and diazotrophic species of bacteria. S. meliloti are motile and possess a cluster of peritrichous flagella. S. meliloti fix atmospheric nitrogen into ammonia for their legume symbionts, such as alfalfa. S. meliloti forms a symbiotic relationship with legumes from the genera Medicago, Melilotus and Trigonella, including the model legume Medicago truncatula. This symbiosis promotes the development of a plant organ, termed a root nodule. Because soil often contains a limited amount of nitrogen for plant use, the symbiotic relationship between S. meliloti and their legume hosts has agricultural applications. These techniques reduce the need for inorganic nitrogenous fertilizers.

Root nodules are found on the roots of plants, primarily legumes, that form a symbiosis with nitrogen-fixing bacteria. Under nitrogen-limiting conditions, capable plants form a symbiotic relationship with a host-specific strain of bacteria known as rhizobia. This process has evolved multiple times within the legumes, as well as in other species found within the Rosid clade. Legume crops include beans, peas, and soybeans.
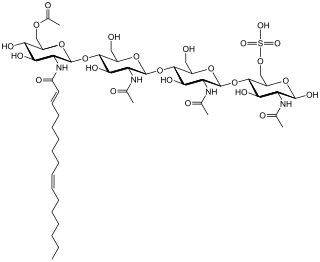
Nod factors, are signaling molecules produced by soil bacteria known as rhizobia in response to flavonoid exudation from plants under nitrogen limited conditions. Nod factors initiate the establishment of a symbiotic relationship between legumes and rhizobia by inducing nodulation. Nod factors produce the differentiation of plant tissue in root hairs into nodules where the bacteria reside and are able to fix nitrogen from the atmosphere for the plant in exchange for photosynthates and the appropriate environment for nitrogen fixation. One of the most important features provided by the plant in this symbiosis is the production of leghemoglobin, which maintains the oxygen concentration low and prevents the inhibition of nitrogenase activity.

The Hyphomicrobiales are an order of Gram-negative Alphaproteobacteria.

Bradyrhizobium is a genus of Gram-negative soil bacteria, many of which fix nitrogen. Nitrogen fixation is an important part of the nitrogen cycle. Plants cannot use atmospheric nitrogen (N2); they must use nitrogen compounds such as nitrates.
Ensifer is a genus of nitrogen-fixing bacteria (rhizobia), three of which have been sequenced.
The nif genes are genes encoding enzymes involved in the fixation of atmospheric nitrogen into a form of nitrogen available to living organisms. The primary enzyme encoded by the nif genes is the nitrogenase complex which is in charge of converting atmospheric nitrogen (N2) to other nitrogen forms such as ammonia which the organism can use for various purposes. Besides the nitrogenase enzyme, the nif genes also encode a number of regulatory proteins involved in nitrogen fixation. The nif genes are found in both free-living nitrogen-fixing bacteria and in symbiotic bacteria associated with various plants. The expression of the nif genes is induced as a response to low concentrations of fixed nitrogen and oxygen concentrations (the low oxygen concentrations are actively maintained in the root environment of host plants). The first Rhizobium genes for nitrogen fixation (nif) and for nodulation (nod) were cloned in the early 1980s by Gary Ruvkun and Sharon R. Long in Frederick M. Ausubel's laboratory.
Funk Brothers Seed Co. v. Kalo Inoculant Co., 333 U.S. 127 (1948), is a United States Supreme Court decision in which the Court held that a facially trivial implementation of a natural principle or phenomenon of nature is not eligible for a patent.
Trophic mutualism is a key type of ecological mutualism. Specifically, "trophic mutualism" refers to the transfer of energy and nutrients between two species. This is also sometimes known as resource-to-resource mutualism. Trophic mutualism often occurs between an autotroph and a heterotroph. Although there are many examples of trophic mutualisms, the heterotroph is generally a fungus or bacteria. This mutualism can be both obligate and opportunistic.

A biofertilizer is a substance which contains living micro-organisms which, when applied to seeds, plant surfaces, or soil, colonize the rhizosphere or the interior of the plant and promotes growth by increasing the supply or availability of primary nutrients to the host plant. Biofertilizers add nutrients through the natural processes of nitrogen fixation, solubilizing phosphorus, and stimulating plant growth through the synthesis of growth-promoting substances. The microorganisms in biofertilizers restore the soil's natural nutrient cycle and build soil organic matter. Through the use of biofertilizers, healthy plants can be grown, while enhancing the sustainability and the health of the soil. Biofertilizers can be expected to reduce the use of synthetic fertilizers and pesticides, but they are not yet able to replace their use. Since they play several roles, a preferred scientific term for such beneficial bacteria is "plant-growth promoting rhizobacteria" (PGPR).
Bradyrhizobium elkanii is a species of legume-root nodulating, microsymbiotic nitrogen-fixing bacterium originally identified as DNA homology group II strains of B. japonicum . In 1988, it was discovered that only DNA homology group II strains caused a destructive bleaching of leaves, termed scientifically "microsymbiont-induced foliar chlorosis", which was widespread in soybean production fields of the southern United States . Whole cell fatty acid content together with antibiotic resistance profiles were major phenotypic differences that helped establish DNA homology group II strains as a new species, Bradyrhizobium elkanii .
Bradyrhizobium yuanmingense is a species of legume-root nodulating, endosymbiont nitrogen-fixing bacterium, associated with Lespedeza and Vigna species. Its type strain is CCBAU 10071(T).
Mesorhizobium mediterraneum is a bacterium from the genus Mesorhizobium, which was isolated from root nodule of the Chickpea in Spain. The species Rhizobium mediterraneum was subsequently transferred to Mesorhizobium mediterraneum. This species, along with many other closely related taxa, have been found to promote production of chickpea and other crops worldwide by forming symbiotic relationships.
Ensifer medicae is a species of gram-negative, nitrogen-fixing, rod-shaped bacteria. They can be free-living or symbionts of leguminous plants in root nodules. E.medicae was first isolated from root nodules on plants in the genus Medicago. Some strains of E.medicae, like WSM419, are aerobic. They are chemoorganotrophic mesophiles that prefer temperatures around 28 °C. In addition to their primary genome, these organisms also have three known plasmids, sized 1,570,951 bp, 1,245,408 bp and 219,313 bp.
Martin Parniske is a German biologist with a specialisation in genetics, microbiology and biochemistry. He is university professor and head of the Institute of Genetics at the Faculty of Biology of the Ludwig Maximilian University of Munich. Parniske's scientific focus is on the molecular interaction between plants and symbiotic and pathogenic organisms including bacteria, fungi, oomycetes and insects.

A symbiosome is a specialised compartment in a host cell that houses an endosymbiont in a symbiotic relationship.










