| Terrabacteria | |
|---|---|
 | |
| Scanning electron micrograph of Actinomyces israelii (Actinomycetota) | |
| Scientific classification | |
| Domain: | Bacteria |
| Clade: | Terrabacteria Battistuzzi et al., 2004, Battistuzzi & Hedges, 2009 |
| Phyla | |
| Synonyms | |
| |
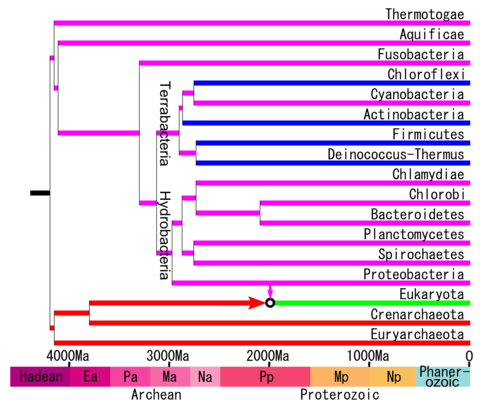
Terrabacteria, also known as kingdom Bacillati, is a taxon containing approximately two-thirds of prokaryote species, including those in the gram positive phyla (Actinomycetota and Bacillota) as well as the phyla "Cyanobacteria", Chloroflexota, and Deinococcota. [1] [2]
It derives its name (terra = "land") from the evolutionary pressures of life on land. Terrabacteria possess important adaptations such as resistance to environmental hazards (e.g., desiccation, ultraviolet radiation, and high salinity) and oxygenic photosynthesis. Also, the unique properties of the cell wall in gram-positive taxa, which likely evolved in response to terrestrial conditions, have contributed toward pathogenicity in many species. [2] These results now leave open the possibility that terrestrial adaptations may have played a larger role in prokaryote evolution than currently understood. [1] [2]
Terrabacteria was proposed in 2004 for Actinomycetota, "Cyanobacteria", and Deinococcota [1] and was expanded later to include Bacillota and Chloroflexota. [2] Other phylogenetic analyses [3] [4] [5] have supported the close relationships of these phyla. Most species of prokaryotes not placed in Terrabacteria were assigned to the taxon Hydrobacteria, [2] (which is also known as kingdom Pseudomonadati) [6] in reference to the moist environment inferred for the common ancestor of those species. Some molecular phylogenetic analyses [7] [8] have not supported this dichotomy of Terrabacteria and Hydrobacteria, but the most recent genomic analyses, [4] [5] including those that have focused on rooting the tree, [4] have found these two groups to be monophyletic. [4]
Terrabacteria and Hydrobacteria were inferred to have diverged approximately 3 billion years ago, suggesting that land (continents) had been colonized by prokaryotes at that time. [2] Together, Terrabacteria and Hydrobacteria form a large group containing 97% of prokaryotes and 99% of all species of Bacteria known by 2009, and placed in the taxon Selabacteria, in allusion to their phototrophic abilities (selas = light). [9] Currently, the bacterial phyla that are outside of Terrabacteria + Hydrobacteria, and thus justifying the taxon Selabacteria, are debated and may or may not include Fusobacteria. [2] [4]
The name “Glidobacteria” [10] included some members of Terrabacteria but excluded the large gram positive groups, Bacillota and Actinomycetota, and is not supported by molecular phylogenetic data. [1] [2] [3] [7] [8] [4] [5] Moreover, the article naming Glidobacteria [10] did not include a molecular phylogeny or statistical analyses and did not follow the widely used three-domain system. For example, it claimed that eukaryotes split from Archaea very recently (~900 Mya), which is contradicted by the fossil record, [11] and that lineage of eukaryotes + Archaea was nested within Bacteria as a close relative of Actinomycetota.
In 2022, new rules were introduced for kingdom-level taxa of prokaryotes, and the same two authors who proposed those new rules, proposed new names in 2024. [12] They concluded that “the taxonomically preferable solution for bacterial kingdoms seems to be to accept the subdivision apparent in the study by Battistuzzi and Hedges,” with refinement. [2] The new (and only valid) name is kingdom Bacillati. [12]