
Nucleotides are organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules within all life-forms on Earth. Nucleotides are obtained in the diet and are also synthesized from common nutrients by the liver.
The urea cycle (also known as the ornithine cycle) is a cycle of biochemical reactions that produces urea (NH2)2CO from ammonia (NH3). Animals that use this cycle, mainly amphibians and mammals, are called ureotelic.

Carbamoyl phosphate is an anion of biochemical significance. In land-dwelling animals, it is an intermediary metabolite in nitrogen disposal through the urea cycle and the synthesis of pyrimidines. Its enzymatic counterpart, carbamoyl phosphate synthetase I, interacts with a class of molecules called sirtuins, NAD dependent protein deacetylases, and ATP to form carbamoyl phosphate. CP then enters the urea cycle in which it reacts with ornithine to form citrulline.

In organic chemistry, a carbamate is a category of organic compounds with the general formula R2NC(O)OR and structure >N−C(=O)−O−, which are formally derived from carbamic acid. The term includes organic compounds, formally obtained by replacing one or more of the hydrogen atoms by other organic functional groups; as well as salts with the carbamate anion H2NCOO−.
Glutamine synthetase (GS) is an enzyme that plays an essential role in the metabolism of nitrogen by catalyzing the condensation of glutamate and ammonia to form glutamine:

Argininosuccinate synthase or synthetase is an enzyme that catalyzes the synthesis of argininosuccinate from citrulline and aspartate. In humans, argininosuccinate synthase is encoded by the ASS gene located on chromosome 9.
Carbamoyl phosphate synthetase I is a ligase enzyme located in the mitochondria involved in the production of urea. Carbamoyl phosphate synthetase I transfers an ammonia molecule to a molecule of bicarbonate that has been phosphorylated by a molecule of ATP. The resulting carbamate is then phosphorylated with another molecule of ATP. The resulting molecule of carbamoyl phosphate leaves the enzyme.
Carbamic acid, which might also be called aminoformic acid or aminocarboxylic acid, is the chemical compound with the formula H2NCOOH. It can be obtained by the reaction of ammonia NH3 and carbon dioxide CO2 at very low temperatures, which also yields ammonium carbamate [NH4]+[NH2CO2]−. The compound is stable only up to about 250 K (−23 °C); at higher temperatures it decomposes into those two gases. The solid apparently consists of dimers, with the two molecules connected by hydrogen bonds between the two carboxyl groups –COOH.

CAD protein is a trifunctional multi-domain enzyme involved in the first three steps of pyrimidine biosynthesis. De-novo synthesis starts with cytosolic carbamoylphosphate synthetase II which uses glutamine, carbon dioxide and ATP. This enzyme is inhibited by uridine triphosphate.
Carbamoyl phosphate synthetase (glutamine-hydrolysing) is an enzyme that catalyzes the reactions that produce carbamoyl phosphate in the cytosol. Its systemic name is hydrogen-carbonate:L-glutamine amido-ligase .

Amidophosphoribosyltransferase (ATase), also known as glutamine phosphoribosylpyrophosphate amidotransferase (GPAT), is an enzyme responsible for catalyzing the conversion of 5-phosphoribosyl-1-pyrophosphate (PRPP) into 5-phosphoribosyl-1-amine (PRA), using the amine group from a glutamine side-chain. This is the committing step in de novo purine synthesis. In humans it is encoded by the PPAT gene. ATase is a member of the purine/pyrimidine phosphoribosyltransferase family.
The enzyme Phosphoribosylaminoimidazole carboxylase, or AIR carboxylase (EC 4.1.1.21) is involved in nucleotide biosynthesis and in particular in purine biosynthesis. It catalyzes the conversion of 5'-phosphoribosyl-5-aminoimidazole ("AIR") into 5'-phosphoribosyl-4-carboxy-5-aminoimidazole ("CAIR") as described in the reaction:

In enzymology, a formate—tetrahydrofolate ligase is an enzyme that catalyzes the chemical reaction
In enzymology, a carbamate kinase (EC 2.7.2.2) is an enzyme that catalyzes the chemical reaction

In molecular biology, glutamine amidotransferases (GATase) are enzymes which catalyse the removal of the ammonia group from a glutamine molecule and its subsequent transfer to a specific substrate, thus creating a new carbon-nitrogen group on the substrate. This activity is found in a range of biosynthetic enzymes, including glutamine amidotransferase, anthranilate synthase component II, p-aminobenzoate, and glutamine-dependent carbamoyl-transferase (CPSase). Glutamine amidotransferase (GATase) domains can occur either as single polypeptides, as in glutamine amidotransferases, or as domains in a much larger multifunctional synthase protein, such as CPSase. On the basis of sequence similarities two classes of GATase domains have been identified: class-I and class-II. Class-I GATase domains are defined by a conserved catalytic triad consisting of cysteine, histidine and glutamate. Class-I GATase domains have been found in the following enzymes: the second component of anthranilate synthase and 4-amino-4-deoxychorismate (ADC) synthase; CTP synthase; GMP synthase; glutamine-dependent carbamoyl-phosphate synthase; phosphoribosylformylglycinamidine synthase II; and the histidine amidotransferase hisH.
In molecular biology, the ATCase/OTCase family is a protein family which contains two related enzymes: aspartate carbamoyltransferase EC 2.1.3.2 and ornithine carbamoyltransferase EC 2.1.3.3. It has been shown that these enzymes are evolutionary related. The predicted secondary structure of both enzymes is similar and there are some regions of sequence similarities. One of these regions includes three residues which have been shown, by crystallographic studies to be implicated in binding the phosphoryl group of carbamoyl phosphate and may also play a role in trimerisation of the molecules. The N-terminal domain is the carbamoyl phosphate binding domain. The C-terminal domain is an aspartate/ornithine-binding domain.
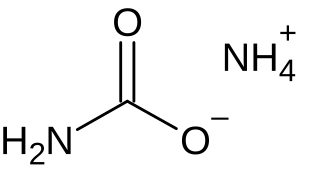
Ammonium carbamate is a chemical compound with the formula [NH4][H2NCO2] consisting of ammonium cation NH+4 and carbamate anion NH2COO−. It is a white solid that is extremely soluble in water, less so in alcohol. Ammonium carbamate can be formed by the reaction of ammonia NH3 with carbon dioxide CO2, and will slowly decompose to those gases at ordinary temperatures and pressures. It is an intermediate in the industrial synthesis of urea (NH2)2CO, an important fertilizer.
Cobyrinate a,c-diamide synthase (EC ), cobyrinic acid a,c-diamide synthetase, CbiA (gene)) is an enzyme which catalyses the chemical reaction
Carbamoyl phosphate synthetase III is one of the three isoforms of the carbamoyl phosphate synthetase, an enzyme that catalyzes the active production of carbamoyl phosphate in many organisms.

In molecular biology, the ATP-grasp fold is a unique ATP-binding protein structural motif made of two α+β subdomains that "grasp" a molecule of ATP between them. ATP-grasp proteins have ATP-dependent carboxylate-amine/thiol ligase activity.

















