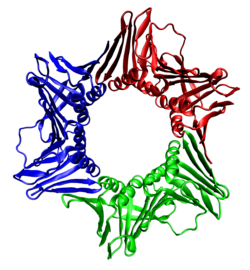

In biochemistry, a protein trimer is a macromolecular complex formed by three, usually non-covalently bound, macromolecules like proteins or nucleic acids. A protein trimer often occurs from the assembly of a protein's quaternary structure. [1] The non-covalent interactions between the hydrophobic and hydrophilic regions on the polypeptides units help to stabilize the quaternary structure. Since a protein trimer is composed of multiple polypeptide subunits, it is considered an oligomer. [2]
Contents
A homotrimer would be formed by three identical molecules. [3] A heterotrimer would be formed by three different macromolecules. Type II Collagen is an example of homotrimeric protein, while Type I collagen is an AAB-type heterotrimeric protein. An example of viral protein homotrimeric protein is mammarenavirus of Z matrix protein. [4]
Porins usually arrange themselves in membranes as trimers.