
Bacillus is a genus of Gram-positive, rod-shaped bacteria, a member of the phylum Bacillota, with 266 named species. The term is also used to describe the shape (rod) of other so-shaped bacteria; and the plural Bacilli is the name of the class of bacteria to which this genus belongs. Bacillus species can be either obligate aerobes which are dependent on oxygen, or facultative anaerobes which can survive in the absence of oxygen. Cultured Bacillus species test positive for the enzyme catalase if oxygen has been used or is present.

An endospore is a dormant, tough, and non-reproductive structure produced by some bacteria in the phylum Bacillota. The name "endospore" is suggestive of a spore or seed-like form, but it is not a true spore. It is a stripped-down, dormant form to which the bacterium can reduce itself. Endospore formation is usually triggered by a lack of nutrients, and usually occurs in gram-positive bacteria. In endospore formation, the bacterium divides within its cell wall, and one side then engulfs the other. Endospores enable bacteria to lie dormant for extended periods, even centuries. There are many reports of spores remaining viable over 10,000 years, and revival of spores millions of years old has been claimed. There is one report of viable spores of Bacillus marismortui in salt crystals approximately 250 million years old. When the environment becomes more favorable, the endospore can reactivate itself into a vegetative state. Most types of bacteria cannot change to the endospore form. Examples of bacterial species that can form endospores include Bacillus cereus, Bacillus anthracis, Bacillus thuringiensis, Clostridium botulinum, and Clostridium tetani. Endospore formation is not found among Archaea.

Desiccation is the state of extreme dryness, or the process of extreme drying. A desiccant is a hygroscopic substance that induces or sustains such a state in its local vicinity in a moderately sealed container.

DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA damage, resulting in tens of thousands of individual molecular lesions per cell per day. Many of these lesions cause structural damage to the DNA molecule and can alter or eliminate the cell's ability to transcribe the gene that the affected DNA encodes. Other lesions induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells after it undergoes mitosis. As a consequence, the DNA repair process is constantly active as it responds to damage in the DNA structure. When normal repair processes fail, and when cellular apoptosis does not occur, irreparable DNA damage may occur, including double-strand breaks and DNA crosslinkages. This can eventually lead to malignant tumors, or cancer as per the two hit hypothesis.

Bacillus subtilis, known also as the hay bacillus or grass bacillus, is a Gram-positive, catalase-positive bacterium, found in soil and the gastrointestinal tract of ruminants, humans and marine sponges. As a member of the genus Bacillus, B. subtilis is rod-shaped, and can form a tough, protective endospore, allowing it to tolerate extreme environmental conditions. B. subtilis has historically been classified as an obligate aerobe, though evidence exists that it is a facultative anaerobe. B. subtilis is considered the best studied Gram-positive bacterium and a model organism to study bacterial chromosome replication and cell differentiation. It is one of the bacterial champions in secreted enzyme production and used on an industrial scale by biotechnology companies.
Cyclobutane is a cycloalkane and organic compound with the formula (CH2)4. Cyclobutane is a colourless gas and is commercially available as a liquefied gas. Derivatives of cyclobutane are called cyclobutanes. Cyclobutane itself is of no commercial or biological significance, but more complex derivatives are important in biology and biotechnology.
Photolyases are DNA repair enzymes that repair damage caused by exposure to ultraviolet light. These enzymes require visible light both for their own activation and for the actual DNA repair. The DNA repair mechanism involving photolyases is called photoreactivation. They mainly convert pyrimidine dimers into a normal pair of pyrimidine bases.
The bacterium, despite its simplicity, contains a well-developed cell structure which is responsible for some of its unique biological structures and pathogenicity. Many structural features are unique to bacteria and are not found among archaea or eukaryotes. Because of the simplicity of bacteria relative to larger organisms and the ease with which they can be manipulated experimentally, the cell structure of bacteria has been well studied, revealing many biochemical principles that have been subsequently applied to other organisms.
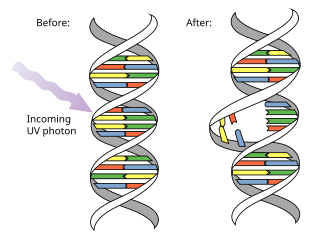
Pyrimidine dimers are molecular lesions formed from thymine or cytosine bases in DNA via photochemical reactions, commonly associated with direct DNA damage. Ultraviolet light induces the formation of covalent linkages between consecutive bases along the nucleotide chain in the vicinity of their carbon–carbon double bonds. The photo-coupled dimers are fluorescent. The dimerization reaction can also occur among pyrimidine bases in dsRNA —uracil or cytosine. Two common UV products are cyclobutane pyrimidine dimers (CPDs) and 6–4 photoproducts. These premutagenic lesions alter the structure of the DNA helix and cause non-canonical base pairing. Specifically, adjacent thymines or cytosines in DNA will form a cyclobutane ring when joined together and cause a distortion in the DNA. This distortion prevents replication or transcription machinery beyond the site of the dimerization. Up to 50–100 such reactions per second might occur in a skin cell during exposure to sunlight, but are usually corrected within seconds by photolyase reactivation or nucleotide excision repair. In humans, the most common form of DNA repair is nucleotide excision repair (NER). In contrast, organisms such as bacteria can counterintuitively harvest energy from the sun to fix DNA damage from pyrimidine dimers via photolyase activity. If these lesions are not fixed, polymerase machinery may misread or add in the incorrect nucleotide to the strand. If the damage to the DNA is overwhelming, mutations can arise within the genome of an organism and may lead to the production of cancer cells. Uncorrected lesions can inhibit polymerases, cause misreading during transcription or replication, or lead to arrest of replication. It causes sunburn and it triggers the production of melanin. Pyrimidine dimers are the primary cause of melanomas in humans.
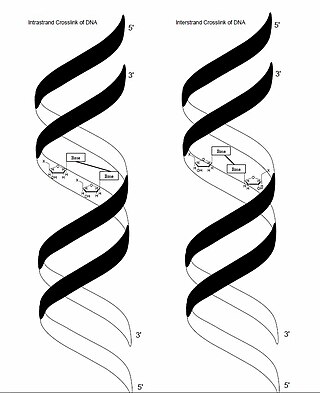
In genetics, crosslinking of DNA occurs when various exogenous or endogenous agents react with two nucleotides of DNA, forming a covalent linkage between them. This crosslink can occur within the same strand (intrastrand) or between opposite strands of double-stranded DNA (interstrand). These adducts interfere with cellular metabolism, such as DNA replication and transcription, triggering cell death. These crosslinks can, however, be repaired through excision or recombination pathways.

Adenylosuccinate lyase is an enzyme that in humans is encoded by the ADSL gene.
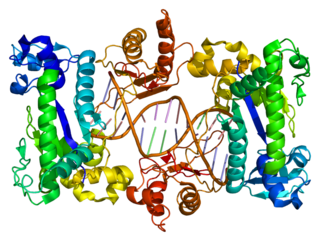
DNA polymerase iota is an enzyme that in humans is encoded by the POLI gene. It is found in higher eukaryotes, and is believed to have arisen from a gene duplication from Pol η. Pol ι, is a Y family polymerase that is involved in translesion synthesis. It can bypass 6-4 pyrimidine adducts and abasic sites and has a high frequency of wrong base incorporation. Like many other Y family polymerases Pol ι, has low processivity, a large DNA binding pocket and doesn't undergo conformational changes when DNA binds. These attributes are what allow Pol ι to carry out its task as a translesion polymerase. Pol ι only uses Hoogsteen base pairing, during DNA synthesis, it will add adenine opposite to thymine in the syn conformation and can add both cytosine and thymine in the anti conformation across guanine, which it flips to the syn conformation.

EXPOSE is a multi-user facility mounted outside the International Space Station (ISS) dedicated to astrobiology. EXPOSE was developed by the European Space Agency (ESA) for long-term spaceflights and was designed to allow exposure of chemical and biological samples to outer space while recording data during exposure.

The FPG IleRS zinc finger domain represents a zinc finger domain found at the C-terminal in both DNA glycosylase/AP lyase enzymes and in isoleucyl tRNA synthetase. In these two types of enzymes, the C-terminal domain forms a zinc finger.

Bacillus subtilis is a rod-shaped, Gram-positive bacteria that is naturally found in soil and vegetation, and is known for its ability to form a small, tough, protective and metabolically dormant endospore. B. subtilis can divide symmetrically to make two daughter cells, or asymmetrically, producing a single endospore that is resistant to environmental factors such as heat, desiccation, radiation and chemical insult which can persist in the environment for long periods of time. The endospore is formed at times of nutritional stress, allowing the organism to persist in the environment until conditions become favourable. The process of endospore formation has profound morphological and physiological consequences: radical post-replicative remodelling of two progeny cells, accompanied eventually by cessation of metabolic activity in one daughter cell and death by lysis of the other.

In molecular biology, the H2TH domain is a DNA-binding domain found in DNA glycosylase/AP lyase enzymes, which are involved in base excision repair of DNA damaged by oxidation or by mutagenic agents. Most damage to bases in DNA is repaired by the base excision repair pathway. These enzymes are primarily from bacteria, and have both DNA glycosylase activity EC 3.2.2.- and AP lyase activity EC 4.2.99.18. Examples include formamidopyrimidine-DNA glycosylases and endonuclease VIII (Nei).
Radical SAM is a designation for a superfamily of enzymes that use a [4Fe-4S]+ cluster to reductively cleave S-adenosyl-L-methionine (SAM) to generate a radical, usually a 5′-deoxyadenosyl radical (5'-dAdo), as a critical intermediate. These enzymes utilize this radical intermediate to perform diverse transformations, often to functionalize unactivated C-H bonds. Radical SAM enzymes are involved in cofactor biosynthesis, enzyme activation, peptide modification, post-transcriptional and post-translational modifications, metalloprotein cluster formation, tRNA modification, lipid metabolism, biosynthesis of antibiotics and natural products etc. The vast majority of known radical SAM enzymes belong to the radical SAM superfamily, and have a cysteine-rich motif that matches or resembles CxxxCxxC. rSAMs comprise the largest superfamily of metal-containing enzymes.
S-specific spore photoproduct lyase is an enzyme with systematic name S-specific spore photoproduct pyrimidine-lyase. This enzyme catalyses the following chemical reaction

Exobiology Radiation Assembly (ERA) was an experiment that investigated the biological effects of space radiation. An astrobiology mission developed by the European Space Agency (ESA), it took place aboard the European Retrievable Carrier (EURECA), an unmanned 4.5 tonne satellite with a payload of 15 experiments.

5,6-Dihydro-5(α-thyminyl)thymine is a DNA pyrimidine dimer photoproduct produced when DNA in bacterial spores is exposed to ultraviolet light. In bacteria, this DNA base dimer deforms the structure of DNA, so endospore forming bacteria have an enzyme called spore photoproduct lyase that repairs this damage.















