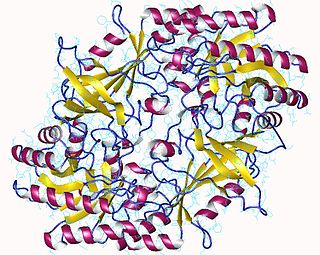
The enzyme ornithine decarboxylase catalyzes the decarboxylation of ornithine to form putrescine. This reaction is the committed step in polyamine synthesis. In humans, this protein has 461 amino acids and forms a homodimer.
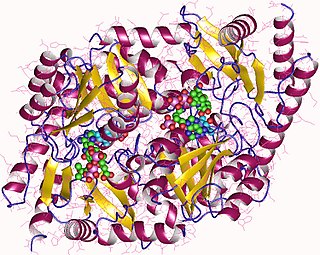
Aminolevulinic acid synthase (ALA synthase, ALAS, or delta-aminolevulinic acid synthase) is an enzyme (EC 2.3.1.37) that catalyzes the synthesis of δ-aminolevulinic acid (ALA) the first common precursor in the biosynthesis of all tetrapyrroles such as hemes, cobalamins and chlorophylls. The reaction is as follows:

Aromatic L-amino acid decarboxylase, also known as DOPA decarboxylase (DDC), tryptophan decarboxylase, and 5-hydroxytryptophan decarboxylase, is a lyase enzyme, located in region 7p12.2-p12.1.

Pyridoxal phosphate (PLP, pyridoxal 5'-phosphate, P5P), the active form of vitamin B6, is a coenzyme in a variety of enzymatic reactions. The International Union of Biochemistry and Molecular Biology has catalogued more than 140 PLP-dependent activities, corresponding to ~4% of all classified activities. The versatility of PLP arises from its ability to covalently bind the substrate, and then to act as an electrophilic catalyst, thereby stabilizing different types of carbanionic reaction intermediates.

Aspartate transaminase (AST) or aspartate aminotransferase, also known as AspAT/ASAT/AAT or (serum) glutamic oxaloacetic transaminase, is a pyridoxal phosphate (PLP)-dependent transaminase enzyme that was first described by Arthur Karmen and colleagues in 1954. AST catalyzes the reversible transfer of an α-amino group between aspartate and glutamate and, as such, is an important enzyme in amino acid metabolism. AST is found in the liver, heart, skeletal muscle, kidneys, brain, red blood cells and gall bladder. Serum AST level, serum ALT level, and their ratio are commonly measured clinically as biomarkers for liver health. The tests are part of blood panels.

Isocitrate dehydrogenase (IDH) (EC 1.1.1.42) and (EC 1.1.1.41) is an enzyme that catalyzes the oxidative decarboxylation of isocitrate, producing alpha-ketoglutarate (α-ketoglutarate) and CO2. This is a two-step process, which involves oxidation of isocitrate (a secondary alcohol) to oxalosuccinate (a ketone), followed by the decarboxylation of the carboxyl group beta to the ketone, forming alpha-ketoglutarate. In humans, IDH exists in three isoforms: IDH3 catalyzes the third step of the citric acid cycle while converting NAD+ to NADH in the mitochondria. The isoforms IDH1 and IDH2 catalyze the same reaction outside the context of the citric acid cycle and use NADP+ as a cofactor instead of NAD+. They localize to the cytosol as well as the mitochondrion and peroxisome.
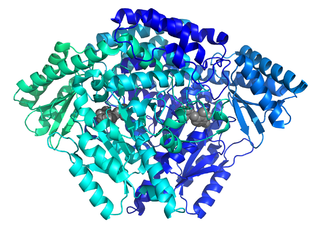
The enzyme histidine decarboxylase is transcribed on chromosome 15, region q21.1-21.2, and catalyzes the decarboxylation of histidine to form histamine. In mammals, histamine is an important biogenic amine with regulatory roles in neurotransmission, gastric acid secretion and immune response. Histidine decarboxylase is the sole member of the histamine synthesis pathway, producing histamine in a one-step reaction. Histamine cannot be generated by any other known enzyme. HDC is therefore the primary source of histamine in most mammals and eukaryotes. The enzyme employs a pyridoxal 5'-phosphate (PLP) cofactor, in similarity to many amino acid decarboxylases. Eukaryotes, as well as gram-negative bacteria share a common HDC, while gram-positive bacteria employ an evolutionarily unrelated pyruvoyl-dependent HDC. In humans, histidine decarboxylase is encoded by the HDC gene.

Oxaloacetate decarboxylase is a carboxy-lyase involved in the conversion of oxaloacetate into pyruvate.

Serine hydroxymethyltransferase (SHMT) is a pyridoxal phosphate (PLP) (Vitamin B6) dependent enzyme (EC 2.1.2.1) which plays an important role in cellular one-carbon pathways by catalyzing the reversible, simultaneous conversions of L-serine to glycine and tetrahydrofolate (THF) to 5,10-methylenetetrahydrofolate (5,10-CH2-THF). This reaction provides the largest part of the one-carbon units available to the cell.
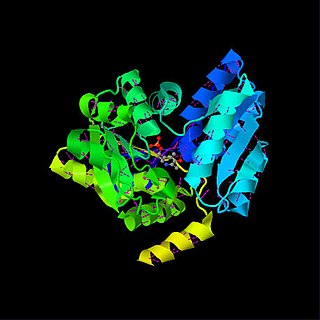
Serine dehydratase or L-serine ammonia lyase (SDH) is in the β-family of pyridoxal phosphate-dependent (PLP) enzymes. SDH is found widely in nature, but its structure and properties vary among species. SDH is found in yeast, bacteria, and the cytoplasm of mammalian hepatocytes. SDH catalyzes the deamination of L-serine to yield pyruvate, with the release of ammonia.

In enzymology, an alanine racemase is an enzyme that catalyzes the chemical reaction
In enzymology, an aspartate racemase is an enzyme that catalyzes the following chemical reaction:

The enzyme aminocyclopropane-1-carboxylic acid synthase catalyzes the synthesis of 1-Aminocyclopropane-1-carboxylic acid (ACC), a precursor for ethylene, from S-Adenosyl methionine, an intermediate in the Yang cycle and activated methyl cycle and a useful molecule for methyl transfer:

Cystathionine beta-lyase, also commonly referred to as CBL or β-cystathionase, is an enzyme that primarily catalyzes the following α,β-elimination reaction

Threonine ammonia-lyase (EC 4.3.1.19, systematic name L-threonine ammonia-lyase (2-oxobutanoate-forming), also commonly referred to as threonine deaminase or threonine dehydratase, is an enzyme responsible for catalyzing the conversion of L-threonine into α-ketobutyrate and ammonia:

The enzyme diaminopimelate decarboxylase (EC 4.1.1.20) catalyzes the cleavage of carbon-carbon bonds in meso 2,6 diaminoheptanedioate to produce CO2 and L-lysine, the essential amino acid. It employs the cofactor pyridoxal phosphate, also known as PLP, which participates in numerous enzymatic transamination, decarboxylation and deamination reactions.

In enzymology, a serine C-palmitoyltransferase (EC 2.3.1.50) is an enzyme that catalyzes the chemical reaction:
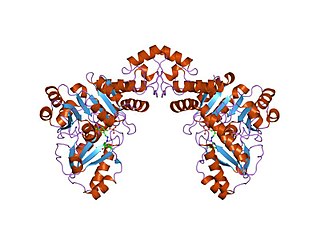
In molecular biology, the Cys/Met metabolism PLP-dependent enzyme family is a family of proteins including enzymes involved in cysteine and methionine metabolism which use PLP (pyridoxal-5'-phosphate) as a cofactor.

In molecular biology, group III pyridoxal-dependent decarboxylases are a family of bacterial enzymes comprising ornithine decarboxylase EC 4.1.1.17, lysine decarboxylase EC 4.1.1.18 and arginine decarboxylase EC 4.1.1.19.

In molecular biology, group IV pyridoxal-dependent decarboxylases are a family of enzymes comprising ornithine decarboxylase EC 4.1.1.17, lysine decarboxylase EC 4.1.1.18, arginine decarboxylase EC 4.1.1.19 and diaminopimelate decarboxylaseEC 4.1.1.20. It is also known as the Orn/Lys/Arg decarboxylase class-II family.






















