Related Research Articles
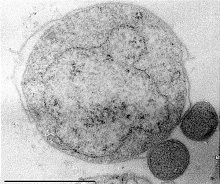
Nanoarchaeum equitans is a species of marine archaea that was discovered in 2002 in a hydrothermal vent off the coast of Iceland on the Kolbeinsey Ridge by Karl Stetter. It has been proposed as the first species in a new phylum, and is the only species within the genus Nanoarchaeum. Strains of this microbe were also found on the Sub-polar Mid Oceanic Ridge, and in the Obsidian Pool in Yellowstone National Park. Since it grows in temperatures approaching boiling, at about 80 °C (176 °F), it is considered to be a thermophile. It grows best in environments with a pH of 6, and a salinity concentration of 2%. Nanoarchaeum appears to be an obligate symbiont on the archaeon Ignicoccus; it must be in contact with the host organism to survive. Nanoarchaeum equitans cannot synthesize lipids but obtains them from its host. Its cells are only 400 nm in diameter, making it the smallest known living organism, and the smallest known archaeon.
Methanogens are anaerobic archaea that produce methane as a byproduct of their energy metabolism, i.e., catabolism. Methane production, or methanogenesis, is the only biochemical pathway for ATP generation in methanogens. All known methanogens belong exclusively to the domain Archaea, although some bacteria, plants, and animal cells are also known to produce methane. However, the biochemical pathway for methane production in these organisms differs from that in methanogens and does not contribute to ATP formation. Methanogens belong to various phyla within the domain Archaea. Previous studies placed all known methanogens into the superphylum Euryarchaeota. However, recent phylogenomic data have led to their reclassification into several different phyla. Methanogens are common in various anoxic environments, such as marine and freshwater sediments, wetlands, the digestive tracts of animals, wastewater treatment plants, rice paddy soil, and landfills. While some methanogens are extremophiles, such as Methanopyrus kandleri, which grows between 84 and 110°C, or Methanonatronarchaeum thermophilum, which grows at a pH range of 8.2 to 10.2 and a Na+ concentration of 3 to 4.8 M, most of the isolates are mesophilic and grow around neutral pH.

The J. Craig Venter Institute (JCVI) is a non-profit genomics research institute founded by J. Craig Venter, Ph.D. in October 2006. The institute was the result of consolidating four organizations: the Center for the Advancement of Genomics, The Institute for Genomic Research (TIGR), the Institute for Biological Energy Alternatives, and the J. Craig Venter Science Foundation Joint Technology Center. It has facilities in Rockville, Maryland, and San Diego, California.
Archaeoglobus is a genus of the phylum Euryarchaeota. Archaeoglobus can be found in high-temperature oil fields where they may contribute to oil field souring.
Methanococcus is a genus of coccoid methanogens of the family Methanococcaceae. They are all mesophiles, except the thermophilic M. thermolithotrophicus and the hyperthermophilic M. jannaschii. The latter was discovered at the base of a “white smoker” chimney at 21°N on the East Pacific Rise and it was the first archaeal genome to be completely sequenced, revealing many novel and eukaryote-like elements.
Methanocaldococcus formerly known as Methanococcus is a genus of coccoid methanogen archaea. They are all mesophiles, except the thermophilic M. thermolithotrophicus and the hyperthermophilic M. jannaschii. The latter was discovered at the base of a “white smoker” chimney at 21°N on the East Pacific Rise and it was the first archaean genome to be completely sequenced, revealing many novel and eukaryote-like elements.
In taxonomy, Thermococcus is a genus of thermophilic Archaea in the family the Thermococcaceae.

Methanobacterium is a genus of the Methanobacteria class in the Archaea kingdom, which produce methane as a metabolic byproduct. Despite the name, this genus belongs not to the bacterial domain but the archaeal domain. Methanobacterium are nonmotile and live without oxygen, which is toxic to them, and they only inhabit anoxic environments.
In enzymology, a coenzyme F420 hydrogenase (EC 1.12.98.1) is an enzyme that catalyzes the chemical reaction
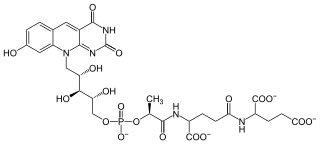
Coenzyme F420 is a family of coenzymes involved in redox reactions in a number of bacteria and archaea. It is derived from coenzyme FO (7,8-didemethyl-8-hydroxy-5-deazariboflavin) and differs by having a oligoglutamyl tail attached via a 2-phospho-L-lactate bridge. F420 is so named because it is a flavin derivative with an absorption maximum at 420 nm.
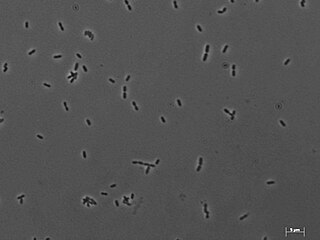
Methanobrevibacter smithii is the predominant methanogenic archaeon in the microbiota of the human gut. M. smithii has a coccobacillus shape. It plays an important role in the efficient digestion of polysaccharides (complex sugars) by consuming the end products of bacterial fermentation (H2, CO2, acetate, and formate). M. smithii is a hydrogenotrophic methanogen that utilizes hydrogen by combining it with carbon dioxide to form methane. The removal of hydrogen by M. smithii is thought to allow an increase in the extraction of energy from nutrients by shifting bacterial fermentation to more oxidized end products.
The archaellum is a unique structure on the cell surface of many archaea that allows for swimming motility. The archaellum consists of a rigid helical filament that is attached to the cell membrane by a molecular motor. This molecular motor – composed of cytosolic, membrane, and pseudo-periplasmic proteins – is responsible for the assembly of the filament and, once assembled, for its rotation. The rotation of the filament propels archaeal cells in liquid medium, in a manner similar to the propeller of a boat. The bacterial analog of the archaellum is the flagellum, which is also responsible for their swimming motility and can also be compared to a rotating corkscrew. Although the movement of archaella and flagella is sometimes described as "whip-like", this is incorrect, as only cilia from Eukaryotes move in this manner. Indeed, even "flagellum" is a misnomer, as bacterial flagella also work as propeller-like structures.

L-2-hydroxycarboxylate dehydrogenase (NAD+) (EC 1.1.1.337, (R)-sulfolactate:NAD+ oxidoreductase, L-sulfolactate dehydrogenase, (R)-sulfolactate dehydrogenase, L-2-hydroxyacid dehydrogenase (NAD+), ComC) is an enzyme with systematic name (2S)-2-hydroxycarboxylate:NAD+ oxidoreductase. This enzyme catalyses the following chemical reaction
Methanococcoides burtonii is a methylotrophic methanogenic archaeon first isolated from Ace Lake, Antarctica. Its type strain is DSM 6242.

Methanococcus maripaludis is a species of methanogenic archaea found in marine environments, predominantly salt marshes. M. maripaludis is a non-pathogenic, gram-negative, weakly motile, non-spore-forming, and strictly anaerobic mesophile. It is classified as a chemolithoautotroph. This archaeon has a pleomorphic coccoid-rod shape of 1.2 by 1.6 μm, in average size, and has many unique metabolic processes that aid in survival. M. maripaludis also has a sequenced genome consisting of around 1.7 Mbp with over 1,700 identified protein-coding genes. In ideal conditions, M. maripaludis grows quickly and can double every two hours.
Methanocaldococcussp. FS406-22 is an archaea in the genus Methanocaldococcus. It is an anaerobic, piezophilic, diazotrophic, hyperthermophilic marine archaeon. This strain is notable for fixing nitrogen at the highest known temperature of nitrogen fixers recorded to date. The 16S rRNA gene of Methanocaldococcus sp. FS406-22, is almost 100% similar to that of Methanocaldococcus jannaschii, a non-nitrogen fixer.

The hydrothermal vent microbial community includes all unicellular organisms that live and reproduce in a chemically distinct area around hydrothermal vents. These include organisms in the microbial mat, free floating cells, or bacteria in an endosymbiotic relationship with animals. Chemolithoautotrophic bacteria derive nutrients and energy from the geological activity at Hydrothermal vents to fix carbon into organic forms. Viruses are also a part of the hydrothermal vent microbial community and their influence on the microbial ecology in these ecosystems is a burgeoning field of research.
5'-deoxyadenosine deaminase is an enzyme that catalyzes the conversion of 5′-deoxyadenosine to 5′-deoxyinosine. To a lesser extent, the enzyme also catalyzes the deamination of 5′-methylthioadenosine, S-adenosylhomocysteine, and adenosine. The molecular mass of the DadD enzyme is approximately 230 kDa. DadD maintains 90% of its enzymatic activity after being heated at 60 degrees Celsius for ten minutes. The preferred pH for 5'deoxyadenosine deaminase is 9.0, with the enzyme denaturing at a pH of 11. The DadD enzyme has a preferred substrate of 5'deoxyadenosine, though it will also react with 5′-methylthioadenosine, S-adenosylhomocysteine, and adenosine at lower efficiencies.
BpsA is a single-module non-ribosomal peptide synthase (NRPS) located in the cytoplasm responsible for the process of creating branched-chain polyamines, and producing spermidine and spermine. It has a singular ligand in its structure involved with Fe3+ and PLIP interactions. As seen by its EC number, it is a transferase (2) that transfers an alkyl or aryl group other than methyl groups (5) (2.5.1). BpsA was first discovered in the archaea Methanococcus jannaschii and thermophile Thermococcus kodakarensis and since then has been used in a variety of applications such as being used as a reporter, researching phosphopantetheinyl transferase (PPTase), and for NRPS domain recombination experiments it can be used as a model. Both (hyper)thermophilic bacteria and euryarchaeotal archaea seem to conserve BpsA and orthologs as branches chains polyamines are crucial for survival. There is also a second type of BpsA also known as Blue-pigment indigoidine synthetase that produces the pigment indigoidine and is found in organisms like Erwinia chrysanthemi. However, not much seems to be known about this variant except that it is a synthase, and it does not yet appear to be classified under an EC number.
References
- ↑ Carol J. Bult; Owen White; Gary J. Olsen; Lixin Zhou; Robert D. Fleischmann; Granger G. Sutton; Judith A. Blake; Lisa M. FitzGerald; Rebecca A. Clayton; Jeannine D. Gocayne; Anthony R. Kerlavage; Brian A. Dougherty; Jean-Francois Tomb; Mark D. Adams; Claudia I. Reich; Ross Overbeek; Ewen F. Kirkness; Keith G. Weinstock; Joseph M. Merrick; Anna Glodek; John L. Scott; Neil S. M. Geoghagen; Janice F. Weidman; Joyce L. Fuhrmann; Dave Nguyen; Teresa R. Utterback; Jenny M. Kelley; Jeremy D. Peterson; Paul W. Sadow; Michael C. Hanna; Matthew D. Cotton; Kevin M. Roberts; Margaret A. Hurst; Brian P. Kaine; Mark Borodovsky; Hans-Peter Klenk; Claire M. Fraser; Hamilton O. Smith; Carl R. Woese; J. Craig Venter (1996). "Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii". Science . 273 (5278): 1058–1073. Bibcode:1996Sci...273.1058B. doi:10.1126/science.273.5278.1058. PMID 8688087. S2CID 41481616.
- 1 2 Robert H. White (2001). "Biosynthesis of the methanogenic cofactors". Vitamins and Hormones . 61: 299–337. doi:10.1016/s0083-6729(01)61010-0. ISBN 978-0-12-709861-6. PMID 11153270.
- 1 2 W. J. Jones; J. A. Leigh; F. Mayer; C. R. Woese; R. S. Wolfe (1983). "Methanococcus jannaschii sp. nov., an extremely thermophilic methanogen from a submarine hydrothermal vent". Archives of Microbiology . 136 (4): 254–261. doi:10.1007/BF00425213. S2CID 33277659.
- ↑ Tsoka, Sophia; Simon, David; Ouzounis, Christos A. (2004). "Automated metabolic reconstruction forMethanococcus jannaschii". Archaea. 1 (4): 223–229. doi: 10.1155/2004/324925 . ISSN 1472-3646. PMC 2685575 . PMID 15810431.
- 1 2 Nicholas Wade (23 August 1996). "Deep sea yields a clue to life's origin". New York Times.
- ↑ Bult, Carol J.; White, Owen; Olsen, Gary J.; Zhou, Lixin; Fleischmann, Robert D.; Sutton, Granger G.; Blake, Judith A.; FitzGerald, Lisa M.; Clayton, Rebecca A.; Gocayne, Jeannine D.; Kerlavage, Anthony R.; Dougherty, Brian A.; Tomb, Jean-Francois; Adams, Mark D.; Reich, Claudia I. (1996-08-23). "Complete Genome Sequence of the Methanogenic Archaeon, Methanococcus jannaschii". Science. 273 (5278): 1058–1073. doi:10.1126/science.273.5278.1058. ISSN 0036-8075.
- ↑ Erica J. Lyon; Seigo Shima; Gerrit Buurman; Shantanu Chowdhuri; Alfred Batschauer; Klaus Steinbach; Rudolf K. Thauer (January 2004). "UV-A/blue-light inactivation of the 'metal-free' hydrogenase (Hmd) from methanogenic archaea". European Journal of Biochemistry . 271 (1): 195–204. doi: 10.1046/j.1432-1033.2003.03920.x . PMID 14686932.
- ↑ Rudolf K. Thauer; Anne-Kristin Kaster; Meike Goenrich; Michael Schick; Takeshi Hiromoto; Seigo Shima (2010). "Hydrogenases from methanogenic archaea, nickel, a novel cofactor, and H2 storage". Annual Review of Biochemistry . 79: 507–536. doi:10.1146/annurev.biochem.030508.152103. PMID 20235826.
- ↑ Wenhong Zhu; Claudia I. Reich; Gary J. Olsen; Carol S. Giometti; John R. Yates III (2004). "Shotgun proteomics of Methanococcus jannaschii and insights into methanogenesis". Journal of Proteome Research . 3 (3): 538–548. doi:10.1021/pr034109s. PMID 15253435.
- ↑ Ilka Haase; Simone Mörtl; Peter Köhler; Adelbert Bacher; Markus Fischer (2003). "Biosynthesis of riboflavin in Archaea: 6,7-dimethyl-8-ribityllumazine synthase of Methanococcus jannaschii". European Journal of Biochemistry . 270 (5): 1025–1032. doi:10.1046/j.1432-1033.2003.03478.x. PMID 12603336.
- ↑ Yoshizumi Ishino; Kayoko Komori; Isaac K. O. Cann; Yosuke Koga (1998). "A novel DNA polymerase family found in Archaea". Journal of Bacteriology . 180 (8): 2232–2236. doi:10.1128/JB.180.8.2232-2236.1998. PMC 107154 . PMID 9555910.
- ↑ Vieille, Claire; Zeikus, Gregory J. (March 2001). "Hyperthermophilic Enzymes: Sources, Uses, and Molecular Mechanisms for Thermostability". Microbiology and Molecular Biology Reviews. 65 (1): 1–43. doi:10.1128/mmbr.65.1.1-43.2001. ISSN 1092-2172. PMC 99017 . PMID 11238984.
- ↑ Tumbula, Debra L.; Whitman, William B. (July 1999). "Genetics of Methanococcus: possibilities for functional genomics in Archaea". Molecular Microbiology. 33 (1): 1–7. doi: 10.1046/j.1365-2958.1999.01463.x . ISSN 0950-382X.
- ↑ Maus, Deborah; Heinz, Jacob; Schirmack, Janosch; Airo, Alessandro; Kounaves, Samuel P.; Wagner, Dirk; Schulze-Makuch, Dirk (2020-01-08). "Methanogenic Archaea Can Produce Methane in Deliquescence-Driven Mars Analog Environments". Scientific Reports. 10 (1): 6. doi:10.1038/s41598-019-56267-4. ISSN 2045-2322. PMC 6949245 . PMID 31913316.