Protein primary structure is the linear sequence of amino acids in a peptide or protein. By convention, the primary structure of a protein is reported starting from the amino-terminal (N) end to the carboxyl-terminal (C) end. Protein biosynthesis is most commonly performed by ribosomes in cells. Peptides can also be synthesized in the laboratory. Protein primary structures can be directly sequenced, or inferred from DNA sequencess.

Proteolysis is the breakdown of proteins into smaller polypeptides or amino acids. Uncatalysed, the hydrolysis of peptide bonds is extremely slow, taking hundreds of years. Proteolysis is typically catalysed by cellular enzymes called proteases, but may also occur by intra-molecular digestion.
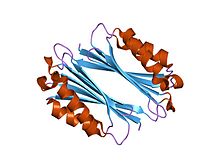
The enzyme ornithine decarboxylase catalyzes the decarboxylation of ornithine to form putrescine. This reaction is the committed step in polyamine synthesis. In humans, this protein has 461 amino acids and forms a homodimer.
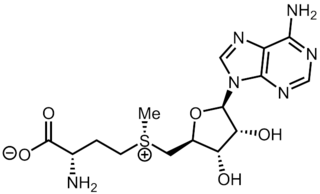
S-Adenosyl methionine (SAM), also known under the commercial names of SAMe, SAM-e, or AdoMet, is a common cosubstrate involved in methyl group transfers, transsulfuration, and aminopropylation. Although these anabolic reactions occur throughout the body, most SAM is produced and consumed in the liver. More than 40 methyl transfers from SAM are known, to various substrates such as nucleic acids, proteins, lipids and secondary metabolites. It is made from adenosine triphosphate (ATP) and methionine by methionine adenosyltransferase. SAM was first discovered by Giulio Cantoni in 1952.

Pyridoxal phosphate (PLP, pyridoxal 5'-phosphate, P5P), the active form of vitamin B6, is a coenzyme in a variety of enzymatic reactions. The International Union of Biochemistry and Molecular Biology has catalogued more than 140 PLP-dependent activities, corresponding to ~4% of all classified activities. The versatility of PLP arises from its ability to covalently bind the substrate, and then to act as an electrophilic catalyst, thereby stabilizing different types of carbanionic reaction intermediates.
Spermine is a polyamine involved in cellular metabolism that is found in all eukaryotic cells. The precursor for synthesis of spermine is the amino acid ornithine. It is an essential growth factor in some bacteria as well. It is found as a polycation at physiological pH. Spermine is associated with nucleic acids and is thought to stabilize helical structure, particularly in viruses. It functions as an intracellular free radical scavenger to protect DNA from free radical attack. Spermine is the chemical primarily responsible for the characteristic odor of semen.

Spermidine is a polyamine compound found in ribosomes and living tissues and having various metabolic functions within organisms. It was originally isolated from semen.

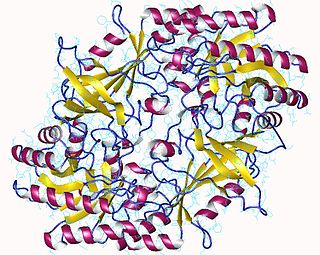
Pyruvate decarboxylase is an enzyme that catalyses the decarboxylation of pyruvic acid to acetaldehyde. It is also called 2-oxo-acid carboxylase, alpha-ketoacid carboxylase, and pyruvic decarboxylase. In anaerobic conditions, this enzyme is participates in the fermentation process that occurs in yeast, especially of the genus Saccharomyces, to produce ethanol by fermentation. It is also present in some species of fish where it permits the fish to perform ethanol fermentation when oxygen is scarce. Pyruvate decarboxylase starts this process by converting pyruvate into acetaldehyde and carbon dioxide. Pyruvate decarboxylase depends on cofactors thiamine pyrophosphate (TPP) and magnesium. This enzyme should not be mistaken for the unrelated enzyme pyruvate dehydrogenase, an oxidoreductase, that catalyzes the oxidative decarboxylation of pyruvate to acetyl-CoA.

Spermidine synthase is an enzyme that catalyzes the transfer of the propylamine group from S-adenosylmethioninamine to putrescine in the biosynthesis of spermidine. The systematic name is S-adenosyl 3-(methylthio)propylamine:putrescine 3-aminopropyltransferase and it belongs to the group of aminopropyl transferases. It does not need any cofactors. Most spermidine synthases exist in solution as dimers.
Hypusine is an uncommon amino acid found in all eukaryotes and in some archaea, but not in bacteria. The only known proteins containing the hypusine residue is eukaryotic translation initiation factor 5A (eIF-5A) and a similar protein found in archaea. In humans, two isoforms of eIF-5A have been described: eIF5A-1 and eIF5A-2. They are encoded by two distinct genes EIF5A and EIF5A2. The protein is involved in protein biosynthesis and promotes the formation of the first peptide bond. The region surrounding the hypusine residue is highly conserved and is essential to the function of eIF5A. Thus, hypusine and eIF-5A appear to be vital for the viability and proliferation of eukaryotic cells.
Spermine synthase is an enzyme that converts spermidine into spermine. This enzyme catalyses the following chemical reaction

Cystathionine beta-lyase, also commonly referred to as CBL or β-cystathionase, is an enzyme that primarily catalyzes the following α,β-elimination reaction

The enzyme Acid-Induced Arginine Decarboxylase (AdiA), also commonly referred to as arginine decarboxylase, catalyzes the conversion of L-arginine into agmatine and carbon dioxide. The process consumes a proton in the decarboxylation and employs a pyridoxal-5'-phosphate (PLP) cofactor, similar to other enzymes involved in amino acid metabolism, such as ornithine decarboxylase and glutamine decarboxylase. It is found in bacteria and virus, though most research has so far focused on forms of the enzyme in bacteria. During the AdiA catalyzed decarboxylation of arginine, the necessary proton is consumed from the cell cytoplasm which helps to prevent the over-accumulation of protons inside the cell and serves to increase the intracellular pH. Arginine decarboxylase is part of an enzymatic system in Escherichia coli, Salmonella Typhimurium, and methane-producing bacteria Methanococcus jannaschii that makes these organisms acid resistant and allows them to survive under highly acidic medium.

The enzyme diaminopimelate decarboxylase (EC 4.1.1.20) catalyzes the cleavage of carbon-carbon bonds in meso 2,6 diaminoheptanedioate to produce CO2 and L-lysine, the essential amino acid. It employs the cofactor pyridoxal phosphate, also known as PLP, which participates in numerous enzymatic transamination, decarboxylation and deamination reactions.
In enzymology, formate C-acetyltransferase is an enzyme. Pyruvate formate lyase is found in Escherichia coli and other organisms. It helps regulate anaerobic glucose metabolism. Using radical non-redox chemistry, it catalyzes the reversible conversion of pyruvate and coenzyme-A into formate and acetyl-CoA. The reaction occurs as follows:

S-Adenosylmethioninamine is a substrate that is required for the biosynthesis of polyamines including spermidine, spermine, and thermospermine. It is produced by decarboxylation of S-adenosyl methionine.

S-Adenosylmethionine synthetase, also known as methionine adenosyltransferase (MAT), is an enzyme that creates S-adenosylmethionine by reacting methionine and ATP.
A polyamine is an organic compound having more than two amino groups. Alkyl polyamines occur naturally, but some are synthetic. Alkylpolyamines are colorless, hygroscopic, and water soluble. Near neutral pH, they exist as the ammonium derivatives. Most aromatic polyamines are crystalline solids at room temperature.
Radical SAM is a designation for a superfamily of enzymes that use a [4Fe-4S]+ cluster to reductively cleave S-adenosyl-L-methionine (SAM) to generate a radical, usually a 5′-deoxyadenosyl radical (5'-dAdo), as a critical intermediate. These enzymes utilize this radical intermediate to perform diverse transformations, often to functionalize unactivated C-H bonds. Radical SAM enzymes are involved in cofactor biosynthesis, enzyme activation, peptide modification, post-transcriptional and post-translational modifications, metalloprotein cluster formation, tRNA modification, lipid metabolism, biosynthesis of antibiotics and natural products etc. The vast majority of known radical SAM enzymes belong to the radical SAM superfamily, and have a cysteine-rich motif that matches or resembles CxxxCxxC. rSAMs comprise the largest superfamily of metal-containing enzymes.
BpsA is a single-module non-ribosomal peptide synthase (NRPS) located in the cytoplasm responsible for the process of creating branched-chain polyamines, and producing spermidine and spermine. It has a singular ligand in its structure involved with Fe3+ and PLIP interactions. As seen by its EC number, it is a transferase (2) that transfers an alkyl or aryl group other than methyl groups (5) (2.5.1). BpsA was first discovered in the archaea Methanococcus jannaschii and thermophile Thermococcus kodakarensis and since then has been used in a variety of applications such as being used as a reporter, researching phosphopantetheinyl transferase (PPTase), and for NRPS domain recombination experiments it can be used as a model. Both (hyper)thermophilic bacteria and euryarchaeotal archaea seem to conserve BpsA and orthologs as branches chains polyamines are crucial for survival. There is also a second type of BpsA also known as Blue-pigment indigoidine synthetase that produces the pigment indigoidine and is found in organisms like Erwinia chrysanthemi. However, not much seems to be known about this variant except that it is a synthase, and it does not yet appear to be classified under an EC number.