A regulatory sequence is a segment of a nucleic acid molecule which is capable of increasing or decreasing the expression of specific genes within an organism. Regulation of gene expression is an essential feature of all living organisms and viruses.

In biology, translation is the process in living cells in which proteins are produced using RNA molecules as templates. The generated protein is a sequence of amino acids. This sequence is determined by the sequence of nucleotides in the RNA. The nucleotides are considered three at a time. Each such triple results in addition of one specific amino acid to the protein being generated. The matching from nucleotide triple to amino acid is called the genetic code. The translation is performed by a large complex of functional RNA and proteins called ribosomes. The entire process is called gene expression.
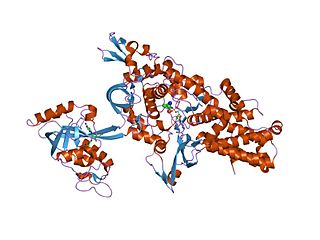
An aminoacyl-tRNA synthetase, also called tRNA-ligase, is an enzyme that attaches the appropriate amino acid onto its corresponding tRNA. It does so by catalyzing the transesterification of a specific cognate amino acid or its precursor to one of all its compatible cognate tRNAs to form an aminoacyl-tRNA. In humans, the 20 different types of aa-tRNA are made by the 20 different aminoacyl-tRNA synthetases, one for each amino acid of the genetic code.
Cis-regulatory elements (CREs) or cis-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphogenesis, the development of anatomy, and other aspects of embryonic development, studied in evolutionary developmental biology.
In genetics, attenuation is a regulatory mechanism for some bacterial operons that results in premature termination of transcription. The canonical example of attenuation used in many introductory genetics textbooks, is ribosome-mediated attenuation of the trp operon. Ribosome-mediated attenuation of the trp operon relies on the fact that, in bacteria, transcription and translation proceed simultaneously. Attenuation involves a provisional stop signal (attenuator), located in the DNA segment that corresponds to the leader sequence of mRNA. During attenuation, the ribosome becomes stalled (delayed) in the attenuator region in the mRNA leader. Depending on the metabolic conditions, the attenuator either stops transcription at that point or allows read-through to the structural gene part of the mRNA and synthesis of the appropriate protein.

Usually found in gram-positive bacteria, the T box leader sequence is an RNA element that controls gene expression through the regulation of translation by binding directly to a specific tRNA and sensing its aminoacylation state. This interaction controls expression of downstream aminoacyl-tRNA synthetase genes, amino acid biosynthesis, and uptake-related genes in a negative feedback loop. The uncharged tRNA acts as the effector for transcription antitermination of genes in the T-box leader family. The anticodon of a specific tRNA base pairs to a specifier sequence within the T-box motif, and the NCCA acceptor tail of the tRNA base pairs to a conserved bulge in the T-box antiterminator hairpin.

The SMKbox riboswitch is an RNA element that regulates gene expression in bacteria. The SMK box riboswitch is found in the 5' UTR of the MetK gene in lactic acid bacteria. The structure of this element changes upon binding to S-adenosyl methionine (SAM) to a conformation that blocks the shine-dalgarno sequence and blocks translation of the gene.

The sucA RNA motif is a conserved RNA structure found in bacteria of the order Burkholderiales. RNAs within this motif are always found in the presumed 5' UTR of sucA genes. sucA encodes a subunit of an enzyme that participates in the citric acid cycle by synthesizing succinyl-CoA from 2-oxoglutarate. A part of the conserved structure overlaps predicted Shine-Dalgarno sequences of the downstream sucA genes. Because of the RNA motif's consistent gene association and a possible mechanism for sequestering the ribosome binding site, it was proposed that the sucA RNA motif corresponds to a cis-regulatory element. Its relatively complex secondary structure could indicate that it is a riboswitch. However, the function of this RNA motif remains unknown.

An RNA thermometer is a temperature-sensitive non-coding RNA molecule which regulates gene expression. Its unique characteristic it is that it does not need proteins or metabolites to function, but only reacts to temperature changes. RNA thermometers often regulate genes required during either a heat shock or cold shock response, but have been implicated in other regulatory roles such as in pathogenicity and starvation.
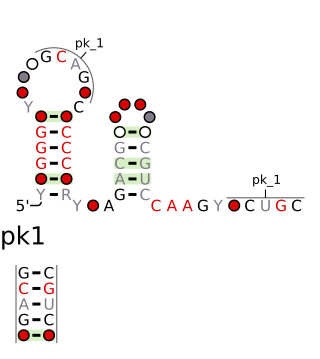
The chrB-a RNA motif and chrB-b RNA motif refer to a related, conserved RNA structure that was discovered by bioinformatics. The structures of these motifs are similar, and some genomic locations are predicted to exhibit both motifs. The chrB-b motif has an extra pseudoknot that is not consistently found in chrB-a examples. It was proposed that the two motifs could be unified into one common structure, with additional information.

The DUF2800 RNA motif is a conserved RNA structure that was discovered by bioinformatics. DUF2800 motif RNAs are found in Bacillota. DUF2800 RNAs are also predicted in the phyla Actinomycetota and Synergistota, although these RNAs are likely the result of recent horizontal gene transfer or conceivably sequence contamination.
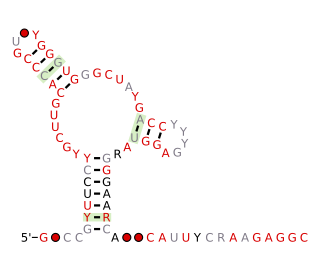
The Fibro-purF RNA motif is a conserved RNA structure that was discovered by bioinformatics. Fibro-purF motif RNAs are found in Fibrobacterota, a group of bacteria that are common in cow rumen. Additionally, the RNAs are found in metagenomic sequences of DNA isolated from cow rumen.

The folE RNA motif, now known as the THF-II riboswitch, is a conserved RNA structure that was discovered by bioinformatics. folE motifs are found in Alphaproteobacteria.
The gltS RNA motif is a conserved RNA structure that was discovered by bioinformatics. gltS motifs are found in the bacterial lineage Vibrionaceae.

The IMES-5 RNA motif is a conserved RNA structure that was discovered by bioinformatics. These RNAs are present in environmental sequences, and have not yet been identified in a classified organism.
lysM RNA motifs are conserved RNA structures that were discovered by bioinformatics. Such bacterial motifs are defined by consistently being upstream of 'lysM' genes, which encode lysin protein domains, a conserved domain that participates in cell wall degradation. lysM motif RNAs likely function as cis-regulatory elements, in view of their positions upstream of protein-coding genes, although this hypothesis is not certain.
malK RNA motifs are conserved RNA structures that were discovered by bioinformatics. They are defined by being consistently located upstream of malK genes, which encode an ATPase that is used by transporters whose ligand is likely a kind of sugar. Most of these genes are annotated either as transporting maltose or glycerol-3-phosphate, however the substrate of the transporters associated with malK motif RNAs has not been experimentally determined. All known types of malK RNA motif are generally located nearby to the Shine-Dalgarno sequence of the downstream gene.

The Mu-like gpT Downstream Element RNA motif is a conserved RNA structure that was discovered by bioinformatics. The Mu-gpT-DE motif is only found in metagenomic sequences arising from unknown organisms.

The osmY RNA motif is a conserved RNA structure that was discovered by bioinformatics. osmY motif RNAs are found in Enterobacteriaceae organisms, although it is not predicted to reside in Escherichia coli.

The Rothia-sucC RNA motif is a conserved RNA structure that was discovered by bioinformatics. Rothia-sucC motif RNAs are found in the actinobacterial genus Rothia.














