
In molecular biology, a riboswitch is a regulatory segment of a messenger RNA molecule that binds a small molecule, resulting in a change in production of the proteins encoded by the mRNA. Thus, an mRNA that contains a riboswitch is directly involved in regulating its own activity, in response to the concentrations of its effector molecule. The discovery that modern organisms use RNA to bind small molecules, and discriminate against closely related analogs, expanded the known natural capabilities of RNA beyond its ability to code for proteins, catalyze reactions, or to bind other RNA or protein macromolecules.

Transfer RNA is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length. In a cell, it provides the physical link between the genetic code in messenger RNA (mRNA) and the amino acid sequence of proteins, carrying the correct sequence of amino acids to be combined by the protein-synthesizing machinery, the ribosome. Each three-nucleotide codon in mRNA is complemented by a three-nucleotide anticodon in tRNA. As such, tRNAs are a necessary component of translation, the biological synthesis of new proteins in accordance with the genetic code.
In genetics, attenuation is a regulatory mechanism for some bacterial operons that results in premature termination of transcription. The canonical example of attenuation used in many introductory genetics textbooks, is ribosome-mediated attenuation of the trp operon. Ribosome-mediated attenuation of the trp operon relies on the fact that, in bacteria, transcription and translation proceed simultaneously. Attenuation involves a provisional stop signal (attenuator), located in the DNA segment that corresponds to the leader sequence of mRNA. During attenuation, the ribosome becomes stalled (delayed) in the attenuator region in the mRNA leader. Depending on the metabolic conditions, the attenuator either stops transcription at that point or allows read-through to the structural gene part of the mRNA and synthesis of the appropriate protein.

The trp operon is a group of genes that are transcribed together, encoding the enzymes that produce the amino acid tryptophan in bacteria. The trp operon was first characterized in Escherichia coli, and it has since been discovered in many other bacteria. The operon is regulated so that, when tryptophan is present in the environment, the genes for tryptophan synthesis are repressed.

The Tryptophan operon leader is an RNA element found at the 5′ of some bacterial tryptophan operons. The leader sequence can form two different structures known as the terminator and the anti-terminator, based on the Tryptophan amounts in the cell. The leader also codes for very short peptide sequence that is rich in tryptophan. The terminator structure is recognised as a termination signal for RNA polymerase and the operon is not transcribed. This structure forms when the cell has an excess of tryptophan and ribosome movement over the leader transcript is not impeded. When there is a deficiency of the charged tryptophanyl tRNA the ribosome translating the leader peptide stalls and the antiterminator structure can form. This allows RNA polymerase to transcribe the operon.

The FMN riboswitch is a highly conserved RNA element which is naturally occurring, and is found frequently in the 5'-untranslated regions of prokaryotic mRNAs that encode for flavin mononucleotide (FMN) biosynthesis and transport proteins. This element is a metabolite-dependent riboswitch that directly binds FMN in the absence of proteins, thus giving it the ability to regulate RNA expression by responding to changes in the concentration of FMN. In Bacillus subtilis, previous studies have shown that this bacterium utilizes at least two FMN riboswitches, where one controls translation initiation, and the other controls premature transcription termination. Regarding the second riboswitch in Bacilius subtilis, premature transcription termination occurs within the 5' untranslated region of the ribDEAHT operon, precluding access to the ribosome-binding site of ypaA mRNA. FMN riboswitches also have various magnesium and potassium ions dispersed throughout the nucleotide structure, some of which participate in binding of FMN.

The Lysine riboswitch is a metabolite binding RNA element found within certain messenger RNAs that serve as a precision sensor for the amino acid lysine. Allosteric rearrangement of mRNA structure is mediated by ligand binding, and this results in modulation of gene expression. Lysine riboswitch are most abundant in Bacillota and Gammaproteobacteria where they are found upstream of a number of genes involved in lysine biosynthesis, transport and catabolism. The lysine riboswitch has also been identified independently and called the L box.
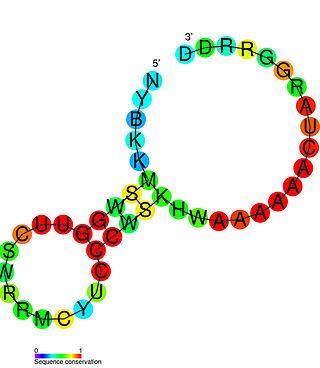
The PreQ1-I riboswitch is a cis-acting element identified in bacteria which regulates expression of genes involved in biosynthesis of the nucleoside queuosine (Q) from GTP. PreQ1 (pre-queuosine1) is an intermediate in the queuosine pathway, and preQ1 riboswitch, as a type of riboswitch, is an RNA element that binds preQ1. The preQ1 riboswitch is distinguished by its unusually small aptamer, compared to other riboswitches. Its atomic-resolution three-dimensional structure has been determined, with the PDB ID 2L1V.

The SAM riboswitch is found upstream of a number of genes which code for proteins involved in methionine or cysteine biosynthesis in Gram-positive bacteria. Two SAM riboswitches in Bacillus subtilis that were experimentally studied act at the level of transcription termination control. The predicted secondary structure consists of a complex stem-loop region followed by a single stem-loop terminator region. An alternative and mutually exclusive form involves bases in the 3' segment of helix 1 with those in the 5' region of helix 5 to form a structure termed the anti-terminator form. When SAM is unbound, the anti-terminator sequence sequesters the terminator sequence so the terminator is unable to form, allowing the polymerase to read-through the downstream gene. When S-Adenosyl methionine (SAM) is bound to the aptamer, the anti-terminator is sequestered by an anti-anti-terminator; the terminator forms and terminates the transcription. However, many SAM riboswitches are likely to regulate gene expression at the level of translation.

The ykkC/yxkD leader is a conserved RNA structure found upstream of the ykkC and yxkD genes in Bacillus subtilis and related genes in other bacteria. The function of this family is unclear for many years although it has been suggested that it may function to switch on efflux pumps and detoxification systems in response to harmful environmental molecules. The Thermoanaerobacter tengcongensis sequence AE013027 overlaps with that of purine riboswitch suggesting that the two riboswitches may work in conjunction to regulate the upstream gene which codes for TTE0584 (Q8RC62), a member of the permease family.

The Ykok leader or M-box is a Mg2+-sensing RNA structure that controls the expression of Magnesium ion transport proteins in bacteria. It is a distinct structure to the Magnesium responsive RNA element.

The SMKbox riboswitch is an RNA element that regulates gene expression in bacteria. The SMK box riboswitch is found in the 5' UTR of the MetK gene in lactic acid bacteria. The structure of this element changes upon binding to S-adenosyl methionine (SAM) to a conformation that blocks the shine-dalgarno sequence and blocks translation of the gene.
The Magnesium responsive RNA element, not to be confused with the completely distinct M-box riboswitch, is a cis-regulatory element that regulates the expression of the magnesium transporter protein MgtA. It is located in the 5' UTR of this gene. The mechanism for the potential magnesium-sensing capacity of this RNA is still unclear, though a recent report suggests that the RNA element targets the mgtA transcript for degradation by RNase E when cells are grown in high Mg2+ environments.

The glutamine riboswitch is a conserved RNA structure that was predicted by bioinformatics. It is present in a variety of lineages of cyanobacteria, as well as some phages that infect cyanobacteria. It is also found in DNA extracted from uncultivated bacteria living in the ocean that are presumably species of cyanobacteria.

The pan RNA motif defines a conserved RNA structure that was identified using bioinformatics. pan motif RNAs are present in three phyla: Chloroflexota, Bacillota, and Pseudomonadota, although within the latter phylum they are only known in deltaproteobacteria. A pan RNA is present in the Firmicute Bacillus subtilis, which is one of the most extensively studied bacteria.

The eps-Associated RNA element is a conserved RNA motif associated with exopolysaccharide (eps) or capsule biosynthesis genes in a subset of bacteria classified within the order Bacillales. It was initially discovered in Bacillus subtilis, located between the second and third gene in the eps operon. Deletion of the EAR element impairs biofilm formation.
Rsa RNAs are non-coding RNAs found in the bacterium Staphylococcus aureus. The shared name comes from their discovery, and does not imply homology. Bioinformatics scans identified the 16 Rsa RNA families named RsaA-K and RsaOA-OG. Others, RsaOH-OX, were found thanks to an RNomic approach. Although the RNAs showed varying expression patterns, many of the newly discovered RNAs were shown to be Hfq-independent and most carried a C-rich motif (UCCC).
In molecular biology, the PyrG leader is a cis-regulatory RNA element found at the 5' of the PyrG mRNA. The PyrG gene encodes a CTP synthase, which is involved in pyrimidine biosynthesis. The PyrG leader regulates expression of PyrG, PyrG can form into two different hairpin structures, a terminator or an anti-terminator. Under low CTP conditions, guanine (G) residues are incorporated at a specific site within the PyrG leader, these allow base-pairing with a uracil (U)-rich region and the formation of an anti-terminator loop, this results in increased expression of PyrG. Under high CTP conditions the guanines are not added, the anti-terminator loop cannot form and instead a terminator loop is formed, preventing further PyrG expression.
LysC is a prokaryotic aspartokinase involved in the biosynthesis of the amino acid lysine. It is found in a variety of bacteria, including Bacillus subtilis, Escherichia coli and Corynebacterium glutamicum. It is notable for containing a riboswitch, a structure in its messenger RNA that prevents its translation when bound to lysine. Such lysine riboswitch thus acts as a mechanism of negative feedback.
An RNA motif is a description of a group of RNAs that have a related structure. RNA motifs consist of a pattern of features within the primary sequence and secondary structure of related RNAs. Thus, it extends the concept of a sequence motif to include RNA secondary structure. The term "RNA motif" can refer both to the pattern and to the RNA sequences that match it.
















