
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.

In molecular genetics, the three prime untranslated region (3′-UTR) is the section of messenger RNA (mRNA) that immediately follows the translation termination codon. The 3′-UTR often contains regulatory regions that post-transcriptionally influence gene expression.
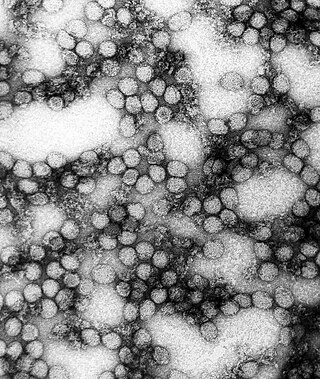
Flavivirus, renamed Orthoflavivirus in 2023, is a genus of positive-strand RNA viruses in the family Flaviviridae. The genus includes the West Nile virus, dengue virus, tick-borne encephalitis virus, yellow fever virus, Zika virus and several other viruses which may cause encephalitis, as well as insect-specific flaviviruses (ISFs) such as cell fusing agent virus (CFAV), Palm Creek virus (PCV), and Parramatta River virus (PaRV). While dual-host flaviviruses can infect vertebrates as well as arthropods, insect-specific flaviviruses are restricted to their competent arthropods. The means by which flaviviruses establish persistent infection in their competent vectors and cause disease in humans depends upon several virus-host interactions, including the intricate interplay between flavivirus-encoded immune antagonists and the host antiviral innate immune effector molecules.

Picornaviruses are a group of related nonenveloped RNA viruses which infect vertebrates including fish, mammals, and birds. They are viruses that represent a large family of small, positive-sense, single-stranded RNA viruses with a 30 nm icosahedral capsid. The viruses in this family can cause a range of diseases including the common cold, poliomyelitis, meningitis, hepatitis, and paralysis.
The 5′ untranslated region is the region of a messenger RNA (mRNA) that is directly upstream from the initiation codon. This region is important for the regulation of translation of a transcript by differing mechanisms in viruses, prokaryotes and eukaryotes. While called untranslated, the 5′ UTR or a portion of it is sometimes translated into a protein product. This product can then regulate the translation of the main coding sequence of the mRNA. In many organisms, however, the 5′ UTR is completely untranslated, instead forming a complex secondary structure to regulate translation.
An internal ribosome entry site, abbreviated IRES, is an RNA element that allows for translation initiation in a cap-independent manner, as part of the greater process of protein synthesis. Initiation of eukaryotic translation nearly always occurs at and is dependent on the 5' cap of mRNA molecules, where the translation initiation complex forms and ribosomes engage the mRNA. IRES elements, however allow ribosomes to engage the mRNA and begin translation independently of the 5' cap.
Ribosome shunting is a mechanism of translation initiation in which ribosomes bypass, or "shunt over", parts of the 5' untranslated region to reach the start codon. However, a benefit of ribosomal shunting is that it can translate backwards allowing more information to be stored than usual in an mRNA molecule. Some viral RNAs have been shown to use ribosome shunting as a more efficient form of translation during certain stages of viral life cycle or when translation initiation factors are scarce. Some viruses known to use this mechanism include adenovirus, Sendai virus, human papillomavirus, duck hepatitis B pararetrovirus, rice tungro bacilliform viruses, and cauliflower mosaic virus. In these viruses the ribosome is directly translocated from the upstream initiation complex to the start codon (AUG) without the need to unwind RNA secondary structures.

A long terminal repeat (LTR) is a pair of identical sequences of DNA, several hundred base pairs long, which occur in eukaryotic genomes on either end of a series of genes or pseudogenes that form a retrotransposon or an endogenous retrovirus or a retroviral provirus. All retroviral genomes are flanked by LTRs, while there are some retrotransposons without LTRs. Typically, an element flanked by a pair of LTRs will encode a reverse transcriptase and an integrase, allowing the element to be copied and inserted at a different location of the genome. Copies of such an LTR-flanked element can often be found hundreds or thousands of times in a genome. LTR retrotransposons comprise about 8% of the human genome.
Luteovirus is a genus of viruses, in the family Tombusviridae. There are 13 species in this genus. Plants serve as natural hosts. The geographical distribution of Luteoviruses is widespread, with the virus primarily infecting plants via transmission by aphid vectors. The virus only replicates within the host cell and not within the vector. The name 'luteovirus' arises from the Latin luteus, which is translated as 'yellow'. Luteovirus was given this name due to the symptomatic yellowing of the plant that occurs as a result of infection.

The Alfalfa mosaic virus RNA 1 5′ UTR stem-loop represents a putative stem-loop structure found in the 5′ UTR in RNA 1 of alfalfa mosaic virus. RNA 1 is responsible for encoding the viral replicase protein P1. This family is required for negative strand RNA synthesis in the alfalfa mosaic virus and may also be involved in positive strand RNA synthesis.
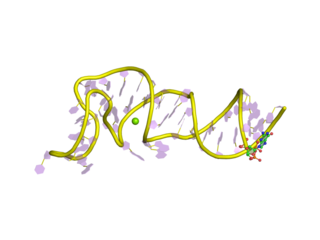
The Coronavirus 3′ stem-loop II-like motif is a secondary structure motif identified in the 3′ untranslated region (3′UTR) of astrovirus, coronavirus and equine rhinovirus genomes. Its function is unknown, but various viral 3′ UTR regions have been found to play roles in viral replication and packaging.
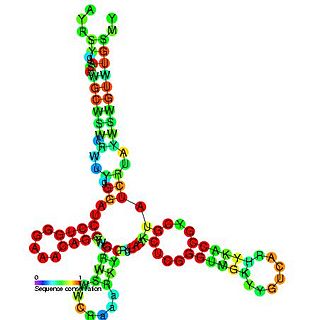
The Barley yellow dwarf virus-like cap-independent translation element (BTE) is an RNA element found in the 3' UTR of some luteoviruses. This element mediates translation of genomic RNA and subgenomic RNA1 (sgRNA1).
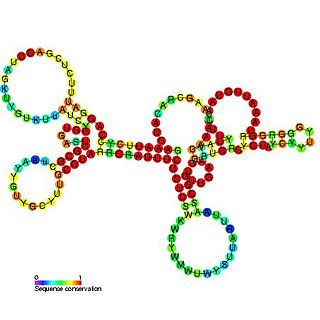
Tombusvirus 5′ UTR is an important cis-regulatory region of the Tombus virus genome.

Red clover necrotic mosaic virus (RCNMV) contains several structural elements present within the 3′ and 5′ untranslated regions (UTR) of the genome that enhance translation. In eukaryotes transcription is a prerequisite for translation. During transcription the pre-mRNA transcript is processes where a 5′ cap is attached onto mRNA and this 5′ cap allows for ribosome assembly onto the mRNA as it acts as a binding site for the eukaryotic initiation factor eIF4F. Once eIF4F is bound to the mRNA this protein complex interacts with the poly(A) binding protein which is present within the 3′ UTR and results in mRNA circularization. This multiprotein-mRNA complex then recruits the ribosome subunits and scans the mRNA until it reaches the start codon. Transcription of viral genomes differs from eukaryotes as viral genomes produce mRNA transcripts that lack a 5’ cap site. Despite lacking a cap site viral genes contain a structural element within the 5’ UTR known as an internal ribosome entry site (IRES). IRES is a structural element that recruits the 40s ribosome subunit to the mRNA within close proximity of the start codon.
In molecular biology, a cap-independent translation element is an RNA sequence found in the 3'UTR of many RNA plant viruses.

In genetics, a kissing stem-loop, or kissing stem loop interaction, is formed in ribonucleic acid (RNA) when two bases between two hairpin loops pair. These intra- and intermolecular kissing interactions are important in forming the tertiary or quaternary structure of many RNAs.
Coronavirus genomes are positive-sense single-stranded RNA molecules with an untranslated region (UTR) at the 5′ end which is called the 5′ UTR. The 5′ UTR is responsible for important biological functions, such as viral replication, transcription and packaging. The 5′ UTR has a conserved RNA secondary structure but different Coronavirus genera have different structural features described below.
Coronavirus genomes are positive-sense single-stranded RNA molecules with an untranslated region (UTR) at the 3′ end which is called the 3′ UTR. The 3′ UTR is responsible for important biological functions, such as viral replication. The 3′ UTR has a conserved RNA secondary structure but different Coronavirus genera have different structural features described below.

Flavivirus 5' UTR are untranslated regions in the genome of viruses in the genus Flavivirus.
Flavivirus 3' UTR are untranslated regions in the genome of viruses in the genus Flavivirus.












