
The National Center for Biotechnology Information (NCBI) is part of the (NLM), a branch of the National Institutes of Health (NIH). It is approved and funded by the government of the United States. The NCBI is located in Bethesda, Maryland, and was founded in 1988 through legislation sponsored by US Congressman Claude Pepper.
A chemical database is a database specifically designed to store chemical information. This information is about chemical and crystal structures, spectra, reactions and syntheses, and thermophysical data.

Content-based image retrieval, also known as query by image content and content-based visual information retrieval (CBVIR), is the application of computer vision techniques to the image retrieval problem, that is, the problem of searching for digital images in large databases. Content-based image retrieval is opposed to traditional concept-based approaches.
BRENDA is the world's most comprehensive online database for functional, biochemical and molecular biological data on enzymes, metabolites and metabolic pathways. It contains data on the properties, function and significance of all enzymes classified by the Enzyme Commission of the International Union of Biochemistry and Molecular Biology (IUBMB). As ELIXIR Core Data Resource and Global Core Biodata Resource, BRENDA is considered a data resource of critical importance to the international life sciences research community. The database compiles a representative overview of enzymes and metabolites using current research data from primary scientific literature and thus serves the purpose of facilitating information retrieval for researchers. BRENDA is subject to the terms of the Creative Commons license, is accessible worldwide and can be used free of charge. As one of the digital resources of the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures, BRENDA is part of the integrated biodata infrastructure DSMZ Digital Diversity.

TAS2R16 is a bitter taste receptor and one of the 25 TAS2Rs. TAS2Rs are receptors that belong to the G-protein-coupled receptors (GPCRs) family. These receptors detect various bitter substances found in nature as agonists, and get stimulated. TAS2R16 receptor is mainly expressed within taste buds present on the surface of the tongue and palate epithelium. TAS2R16 is activated by bitter β-glucopyranosides
A receptor activated solely by a synthetic ligand (RASSL) or designer receptor exclusively activated by designer drugs (DREADD), is a class of artificially engineered protein receptors used in the field of chemogenetics which are selectively activated by certain ligands. They are used in biomedical research, in particular in neuroscience to manipulate the activity of neurons.
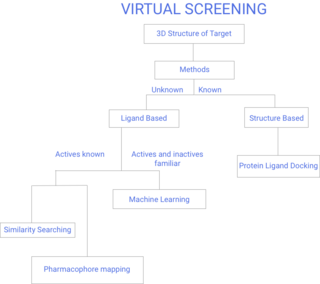
Virtual screening (VS) is a computational technique used in drug discovery to search libraries of small molecules in order to identify those structures which are most likely to bind to a drug target, typically a protein receptor or enzyme.
ChemSpider is a freely accessible online database of chemicals owned by the Royal Society of Chemistry. It contains information on more than 100 million molecules from over 270 data sources, each of them receiving a unique identifier called ChemSpider Identifier.
BLAT is a pairwise sequence alignment algorithm that was developed by Jim Kent at the University of California Santa Cruz (UCSC) in the early 2000s to assist in the assembly and annotation of the human genome. It was designed primarily to decrease the time needed to align millions of mouse genomic reads and expressed sequence tags against the human genome sequence. The alignment tools of the time were not capable of performing these operations in a manner that would allow a regular update of the human genome assembly. Compared to pre-existing tools, BLAT was ~500 times faster with performing mRNA/DNA alignments and ~50 times faster with protein/protein alignments.

A taste receptor or tastant is a type of cellular receptor that facilitates the sensation of taste. When food or other substances enter the mouth, molecules interact with saliva and are bound to taste receptors in the oral cavity and other locations. Molecules which give a sensation of taste are considered "sapid".

Taste receptor 2 member 38 is a protein that in humans is encoded by the TAS2R38 gene. TAS2R38 is a bitter taste receptor; varying genotypes of TAS2R38 influence the ability to taste both 6-n-propylthiouracil (PROP) and phenylthiocarbamide (PTC). Though it has often been proposed that varying taste receptor genotypes could influence tasting ability, TAS2R38 is one of the few taste receptors shown to have this function.
Gustducin is a G protein associated with taste and the gustatory system, found in some taste receptor cells. Research on the discovery and isolation of gustducin is recent. It is known to play a large role in the transduction of bitter, sweet and umami stimuli. Its pathways are many and diverse.

Taste receptor type 2 member 1 (TAS2R1/T2R1) is a protein that in humans is encoded by the TAS2R1 gene. It belongs to the G protein-coupled receptor (GPCR) family and is related to class A-like GPCRs, they contain 7 transmembrane helix bundles and short N-terminus loop. Furthermore, TAS2R1 is member of the 25 known human bitter taste receptors, which enable the perception of bitter taste in the mouth cavity. Increasing evidence indicates a functional role of TAS2Rs in extra-oral tissues.

Taste receptor type 2 member 10 is a protein that in humans is encoded by the TAS2R10 gene. The protein is responsible for bitter taste recognition in mammals. It serves as a defense mechanism to prevent consumption of toxic substances which often have a characteristic bitter taste.

Taste receptor type 2 member 14 is a protein that in humans is encoded by the TAS2R14 gene.

Taste receptor type 1 member 1 is a protein that in humans is encoded by the TAS1R1 gene.

Taste receptors for bitter substances (T2Rs/TAS2Rs) belong to the family of G-protein coupled receptors and are related to class A-like GPCRs. There are 25 known T2Rs in humans responsible for bitter taste perception.
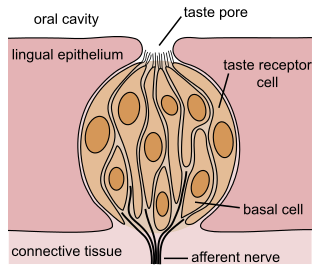
The gustatory system or sense of taste is the sensory system that is partially responsible for the perception of taste (flavor). Taste is the perception stimulated when a substance in the mouth reacts chemically with taste receptor cells located on taste buds in the oral cavity, mostly on the tongue. Taste, along with the sense of smell and trigeminal nerve stimulation, determines flavors of food and other substances. Humans have taste receptors on taste buds and other areas, including the upper surface of the tongue and the epiglottis. The gustatory cortex is responsible for the perception of taste.
GeneCards is a database of human genes that provides genomic, proteomic, transcriptomic, genetic and functional information on all known and predicted human genes. It is being developed and maintained by the Crown Human Genome Center at the Weizmann Institute of Science, in collaboration with LifeMap Sciences.
The E. coli Metabolome Database (ECMDB) is a freely accessible, online database of small molecule metabolites found in or produced by Escherichia coli. Escherichia coli is perhaps the best studied bacterium on earth and has served as the "model microbe" in microbiology research for more than 60 years. The ECMDB is essentially an E. coli "omics" encyclopedia containing detailed data on the genome, proteome and metabolome of E. coli. ECMDB is part of a suite of organism-specific metabolomics databases that includes DrugBank, HMDB, YMDB and SMPDB. As a metabolomics resource, the ECMDB is designed to facilitate research in the area gut/microbiome metabolomics and environmental metabolomics. The ECMDB contains two kinds of data: 1) chemical data and 2) molecular biology and/or biochemical data. The chemical data includes more than 2700 metabolite structures with detailed metabolite descriptions along with nearly 5000 NMR, GC-MS and LC-MS spectra corresponding to these metabolites. The biochemical data includes nearly 1600 protein sequences and more than 3100 biochemical reactions that are linked to these metabolite entries. Each metabolite entry in the ECMDB contains more than 80 data fields with approximately 65% of the information being devoted to chemical data and the other 35% of the information devoted to enzymatic or biochemical data. Many data fields are hyperlinked to other databases. The ECMDB also has a variety of structure and pathway viewing applets. The ECMDB database offers a number of text, sequence, spectral, chemical structure and relational query searches. These are described in more detail below.










