Related Research Articles
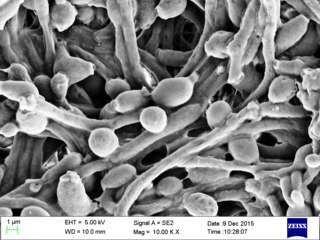
Candida albicans is an opportunistic pathogenic yeast that is a common member of the human gut flora. It can also survive outside the human body. It is detected in the gastrointestinal tract and mouth in 40–60% of healthy adults. It is usually a commensal organism, but it can become pathogenic in immunocompromised individuals under a variety of conditions. It is one of the few species of the genus Candida that cause the human infection candidiasis, which results from an overgrowth of the fungus. Candidiasis is, for example, often observed in HIV-infected patients. C. albicans is the most common fungal species isolated from biofilms either formed on (permanent) implanted medical devices or on human tissue. C. albicans, C. tropicalis, C. parapsilosis, and C. glabrata are together responsible for 50–90% of all cases of candidiasis in humans. A mortality rate of 40% has been reported for patients with systemic candidiasis due to C. albicans. By one estimate, invasive candidiasis contracted in a hospital causes 2,800 to 11,200 deaths yearly in the US. Nevertheless, these numbers may not truly reflect the true extent of damage this organism causes, given new studies indicating that C. albicans can cross the blood–brain barrier in mice.
In genetics, a transcription terminator is a section of nucleic acid sequence that marks the end of a gene or operon in genomic DNA during transcription. This sequence mediates transcriptional termination by providing signals in the newly synthesized transcript RNA that trigger processes which release the transcript RNA from the transcriptional complex. These processes include the direct interaction of the mRNA secondary structure with the complex and/or the indirect activities of recruited termination factors. Release of the transcriptional complex frees RNA polymerase and related transcriptional machinery to begin transcription of new mRNAs.

Lamins, also known as nuclear lamins are fibrous proteins in type V intermediate filaments, providing structural function and transcriptional regulation in the cell nucleus. Nuclear lamins interact with inner nuclear membrane proteins to form the nuclear lamina on the interior of the nuclear envelope. Lamins have elastic and mechanosensitive properties, and can alter gene regulation in a feedback response to mechanical cues. Lamins are present in all animals but are not found in microorganisms, plants or fungi. Lamin proteins are involved in the disassembling and reforming of the nuclear envelope during mitosis, the positioning of nuclear pores, and programmed cell death. Mutations in lamin genes can result in several genetic laminopathies, which may be life-threatening.

Two-hybrid screening is a molecular biology technique used to discover protein–protein interactions (PPIs) and protein–DNA interactions by testing for physical interactions between two proteins or a single protein and a DNA molecule, respectively.
RNA-binding proteins are proteins that bind to the double or single stranded RNA in cells and participate in forming ribonucleoprotein complexes. RBPs contain various structural motifs, such as RNA recognition motif (RRM), dsRNA binding domain, zinc finger and others. They are cytoplasmic and nuclear proteins. However, since most mature RNA is exported from the nucleus relatively quickly, most RBPs in the nucleus exist as complexes of protein and pre-mRNA called heterogeneous ribonucleoprotein particles (hnRNPs). RBPs have crucial roles in various cellular processes such as: cellular function, transport and localization. They especially play a major role in post-transcriptional control of RNAs, such as: splicing, polyadenylation, mRNA stabilization, mRNA localization and translation. Eukaryotic cells express diverse RBPs with unique RNA-binding activity and protein–protein interaction. According to the Eukaryotic RBP Database (EuRBPDB), there are 2961 genes encoding RBPs in humans. During evolution, the diversity of RBPs greatly increased with the increase in the number of introns. Diversity enabled eukaryotic cells to utilize RNA exons in various arrangements, giving rise to a unique RNP (ribonucleoprotein) for each RNA. Although RBPs have a crucial role in post-transcriptional regulation in gene expression, relatively few RBPs have been studied systematically.It has now become clear that RNA–RBP interactions play important roles in many biological processes among organisms.
Mothers against decapentaplegic homolog 2, also known as SMAD family member 2 or SMAD2, is a protein that in humans is encoded by the SMAD2 gene. MAD homolog 2 belongs to the SMAD, a family of proteins similar to the gene products of the Drosophila gene 'mothers against decapentaplegic' (Mad) and the C. elegans gene Sma. SMAD proteins are signal transducers and transcriptional modulators that mediate multiple signaling pathways.

Protein inhibitor of activated STAT (PIAS), also known as E3 SUMO-protein ligase PIAS, is a protein that regulates transcription in mammals. PIAS proteins act as transcriptional co-regulators with at least 60 different proteins in order to either activate or repress transcription. The transcription factors STAT, NF-κB, p73, and p53 are among the many proteins that PIAS interacts with.

In genetics and molecular biology, a corepressor is a molecule that represses the expression of genes. In prokaryotes, corepressors are small molecules whereas in eukaryotes, corepressors are proteins. A corepressor does not directly bind to DNA, but instead indirectly regulates gene expression by binding to repressors.

The initiator element (Inr), sometimes referred to as initiator motif, is a core promoter that is similar in function to the Pribnow box or the TATA box. The Inr is the simplest functional promoter that is able to direct transcription initiation without a functional TATA box. It has the consensus sequence YYANWYY in humans. Similarly to the TATA box, the Inr element facilitates the binding of transcription Factor II D (TFIID). The Inr works by enhancing binding affinity and strengthening the promoter.
TEAD2, together with TEAD1, defines a novel family of transcription factors, the TEAD family, highly conserved through evolution. TEAD proteins were notably found in Drosophila (Scalloped), C. elegans, S. cerevisiae and A. nidulans. TEAD2 has been less studied than TEAD1 but a few studies revealed its role during development.

Nuclear factor of activated T-cells, cytoplasmic 2 is a protein that in humans is encoded by the NFATC2 gene.

Lymphoid enhancer-binding factor 1 (LEF1) is a protein that in humans is encoded by the LEF1 gene. It is a member of T cell factor/lymphoid enhancer factor (TCF/LEF) family.

Histone deacetylase 4, also known as HDAC4, is a protein that in humans is encoded by the HDAC4 gene.

Transcription factor E2F2 is a protein that in humans is encoded by the E2F2 gene.

YAP1, also known as YAP or YAP65, is a protein that acts as a transcription coregulator that promotes transcription of genes involved in cellular proliferation and suppressing apoptotic genes. YAP1 is a component in the hippo signaling pathway which regulates organ size, regeneration, and tumorigenesis. YAP1 was first identified by virtue of its ability to associate with the SH3 domain of Yes and Src protein tyrosine kinases. YAP1 is a potent oncogene, which is amplified in various human cancers.

Transcriptional regulator Kaiso is a protein that in humans is encoded by the ZBTB33 gene. This gene encodes a transcriptional regulator with bimodal DNA-binding specificity, which binds to methylated CGCG and also to the non-methylated consensus KAISO-binding site TCCTGCNA. The protein contains an N-terminal POZ/BTB domain and 3 C-terminal zinc finger motifs. It recruits the N-CoR repressor complex to promote histone deacetylation and the formation of repressive chromatin structures in target gene promoters. It may contribute to the repression of target genes of the Wnt signaling pathway, and may also activate transcription of a subset of target genes by the recruitment of catenin delta-2 (CTNND2). Its interaction with catenin delta-1 (CTNND1) inhibits binding to both methylated and non-methylated DNA. It also interacts directly with the nuclear import receptor Importin-α2, which may mediate nuclear import of this protein. Alternatively spliced transcript variants encoding the same protein have been identified.

Cyclic AMP-responsive element-binding protein 3 is a protein that in humans is encoded by the CREB3 gene.

DSIF is a protein complex that can either negatively or positively affect transcription by RNA polymerase II. It can interact with the negative elongation factor (NELF) to promote the stalling of Pol II at some genes, which is called promoter proximal pausing. The pause occurs soon after initiation, once 20-60 nucleotides have been transcribed. This stalling is relieved by positive transcription elongation factor b (P-TEFb) and Pol II enters productive elongation to resume synthesis till finish. In humans, DSIF is composed of hSPT4 and hSPT5. hSPT5 has a direct role in mRNA capping which occurs while the elongation is paused.
In molecular biology, the BEN domain is a protein domain which is found in diverse proteins including:
The TREX (TRanscription-EXport) complex is a conserved eukaryotic multi-protein complex that couples mRNA transcription and nuclear export. The TREX complex travels across transcribed genes with RNA polymerase II. TREX binds mRNA and recruits transport proteins NXF1 and NXT1, which shuttle the mRNA out of the nucleus. The TREX complex plays an important role in genome stability and neurodegenerative diseases.
References
- 1 2 Steinbach, William J.; Reedy, Jennifer L.; Cramer, Robert A.; Perfect, John R.; Heitman, Joseph (June 2007). "Harnessing calcineurin as a novel anti-infective agent against invasive fungal infections". Nature Reviews. Microbiology. 5 (6): 418–430. doi:10.1038/nrmicro1680. ISSN 1740-1534. PMID 17505522. S2CID 28292376.
- ↑ Stathopoulos-Gerontides, A.; Guo, J. J.; Cyert, M. S. (1999-04-01). "Yeast calcineurin regulates nuclear localization of the Crz1p transcription factor through dephosphorylation". Genes & Development. 13 (7): 798–803. doi:10.1101/gad.13.7.798. ISSN 0890-9369. PMC 316598 . PMID 10197980.
- 1 2 Stathopoulos, A. M.; Cyert, M. S. (1997-12-15). "Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast". Genes & Development. 11 (24): 3432–3444. doi:10.1101/gad.11.24.3432. ISSN 0890-9369. PMC 316814 . PMID 9407035.
- ↑ Karababa, Mahir; Valentino, Emilio; Pardini, Giacomo; Coste, Alix T.; Bille, Jacques; Sanglard, Dominique (March 2006). "CRZ1 , a target of the calcineurin pathway in Candida albicans: Calcineurin targets in Candida albicans". Molecular Microbiology. 59 (5): 1429–1451. doi: 10.1111/j.1365-2958.2005.05037.x . PMID 16468987.