
Nonlinear dimensionality reduction, also known as manifold learning, is any of various related techniques that aim to project high-dimensional data onto lower-dimensional latent manifolds, with the goal of either visualizing the data in the low-dimensional space, or learning the mapping itself. The techniques described below can be understood as generalizations of linear decomposition methods used for dimensionality reduction, such as singular value decomposition and principal component analysis.

Cluster analysis or clustering is the task of grouping a set of objects in such a way that objects in the same group are more similar to each other than to those in other groups (clusters). It is a main task of exploratory data analysis, and a common technique for statistical data analysis, used in many fields, including pattern recognition, image analysis, information retrieval, bioinformatics, data compression, computer graphics and machine learning.
In mathematics, computer science and especially graph theory, a distance matrix is a square matrix containing the distances, taken pairwise, between the elements of a set. Depending upon the application involved, the distance being used to define this matrix may or may not be a metric. If there are N elements, this matrix will have size N×N. In graph-theoretic applications, the elements are more often referred to as points, nodes or vertices.
The scale-invariant feature transform (SIFT) is a computer vision algorithm to detect, describe, and match local features in images, invented by David Lowe in 1999. Applications include object recognition, robotic mapping and navigation, image stitching, 3D modeling, gesture recognition, video tracking, individual identification of wildlife and match moving.
In statistics, the k-nearest neighbors algorithm (k-NN) is a non-parametric supervised learning method first developed by Evelyn Fix and Joseph Hodges in 1951, and later expanded by Thomas Cover. It is used for classification and regression. In both cases, the input consists of the k closest training examples in a data set. The output depends on whether k-NN is used for classification or regression:
k-means clustering is a method of vector quantization, originally from signal processing, that aims to partition n observations into k clusters in which each observation belongs to the cluster with the nearest mean, serving as a prototype of the cluster. This results in a partitioning of the data space into Voronoi cells. k-means clustering minimizes within-cluster variances, but not regular Euclidean distances, which would be the more difficult Weber problem: the mean optimizes squared errors, whereas only the geometric median minimizes Euclidean distances. For instance, better Euclidean solutions can be found using k-medians and k-medoids.
Medoids are representative objects of a data set or a cluster within a data set whose sum of dissimilarities to all the objects in the cluster is minimal. Medoids are similar in concept to means or centroids, but medoids are always restricted to be members of the data set. Medoids are most commonly used on data when a mean or centroid cannot be defined, such as graphs. They are also used in contexts where the centroid is not representative of the dataset like in images, 3-D trajectories and gene expression. These are also of interest while wanting to find a representative using some distance other than squared euclidean distance.
Fuzzy clustering is a form of clustering in which each data point can belong to more than one cluster.
Nearest neighbor search (NNS), as a form of proximity search, is the optimization problem of finding the point in a given set that is closest to a given point. Closeness is typically expressed in terms of a dissimilarity function: the less similar the objects, the larger the function values.
The study of facility location problems (FLP), also known as location analysis, is a branch of operations research and computational geometry concerned with the optimal placement of facilities to minimize transportation costs while considering factors like avoiding placing hazardous materials near housing, and competitors' facilities. The techniques also apply to cluster analysis.
Density-based spatial clustering of applications with noise (DBSCAN) is a data clustering algorithm proposed by Martin Ester, Hans-Peter Kriegel, Jörg Sander and Xiaowei Xu in 1996. It is a density-based clustering non-parametric algorithm: given a set of points in some space, it groups together points that are closely packed, and marks as outliers points that lie alone in low-density regions . DBSCAN is one of the most common, and most commonly cited, clustering algorithms.
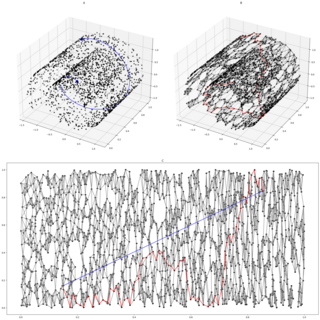
Isomap is a nonlinear dimensionality reduction method. It is one of several widely used low-dimensional embedding methods. Isomap is used for computing a quasi-isometric, low-dimensional embedding of a set of high-dimensional data points. The algorithm provides a simple method for estimating the intrinsic geometry of a data manifold based on a rough estimate of each data point’s neighbors on the manifold. Isomap is highly efficient and generally applicable to a broad range of data sources and dimensionalities.
Consensus clustering is a method of aggregating results from multiple clustering algorithms. Also called cluster ensembles or aggregation of clustering, it refers to the situation in which a number of different (input) clusterings have been obtained for a particular dataset and it is desired to find a single (consensus) clustering which is a better fit in some sense than the existing clusterings. Consensus clustering is thus the problem of reconciling clustering information about the same data set coming from different sources or from different runs of the same algorithm. When cast as an optimization problem, consensus clustering is known as median partition, and has been shown to be NP-complete, even when the number of input clusterings is three. Consensus clustering for unsupervised learning is analogous to ensemble learning in supervised learning.
BIRCH is an unsupervised data mining algorithm used to perform hierarchical clustering over particularly large data-sets. With modifications it can also be used to accelerate k-means clustering and Gaussian mixture modeling with the expectation–maximization algorithm. An advantage of BIRCH is its ability to incrementally and dynamically cluster incoming, multi-dimensional metric data points in an attempt to produce the best quality clustering for a given set of resources. In most cases, BIRCH only requires a single scan of the database.
Silhouette refers to a method of interpretation and validation of consistency within clusters of data. The technique provides a succinct graphical representation of how well each object has been classified. It was proposed by Belgian statistician Peter Rousseeuw in 1987.
Clustering high-dimensional data is the cluster analysis of data with anywhere from a few dozen to many thousands of dimensions. Such high-dimensional spaces of data are often encountered in areas such as medicine, where DNA microarray technology can produce many measurements at once, and the clustering of text documents, where, if a word-frequency vector is used, the number of dimensions equals the size of the vocabulary.
The Davies–Bouldin index (DBI), introduced by David L. Davies and Donald W. Bouldin in 1979, is a metric for evaluating clustering algorithms. This is an internal evaluation scheme, where the validation of how well the clustering has been done is made using quantities and features inherent to the dataset. This has a drawback that a good value reported by this method does not imply the best information retrieval.
In computer science, data stream clustering is defined as the clustering of data that arrive continuously such as telephone records, multimedia data, financial transactions etc. Data stream clustering is usually studied as a streaming algorithm and the objective is, given a sequence of points, to construct a good clustering of the stream, using a small amount of memory and time.
The Dunn index (DI) (introduced by J. C. Dunn in 1974) is a metric for evaluating clustering algorithms. This is part of a group of validity indices including the Davies–Bouldin index or Silhouette index, in that it is an internal evaluation scheme, where the result is based on the clustered data itself. As do all other such indices, the aim is to identify sets of clusters that are compact, with a small variance between members of the cluster, and well separated, where the means of different clusters are sufficiently far apart, as compared to the within cluster variance. For a given assignment of clusters, a higher Dunn index indicates better clustering. One of the drawbacks of using this is the computational cost as the number of clusters and dimensionality of the data increase.
In data mining and machine learning, kq-flats algorithm is an iterative method which aims to partition m observations into k clusters where each cluster is close to a q-flat, where q is a given integer.





