
Lipoic acid (LA), also known as α-lipoic acid, alpha-lipoic acid (ALA) and thioctic acid, is an organosulfur compound derived from caprylic acid (octanoic acid). ALA is made in animals normally, and is essential for aerobic metabolism. It is also manufactured and is available as a dietary supplement in some countries where it is marketed as an antioxidant, and is available as a pharmaceutical drug in other countries. Lipoate is the conjugate base of lipoic acid, and the most prevalent form of LA under physiological conditions. Only the (R)-(+)-enantiomer (RLA) exists in nature and is essential for aerobic metabolism because RLA is an essential cofactor of many enzyme complexes.

Acetyl-CoA carboxylase (ACC) is a biotin-dependent enzyme that catalyzes the irreversible carboxylation of acetyl-CoA to produce malonyl-CoA through its two catalytic activities, biotin carboxylase (BC) and carboxyltransferase (CT). ACC is a multi-subunit enzyme in most prokaryotes and in the chloroplasts of most plants and algae, whereas it is a large, multi-domain enzyme in the cytoplasm of most eukaryotes. The most important function of ACC is to provide the malonyl-CoA substrate for the biosynthesis of fatty acids. The activity of ACC can be controlled at the transcriptional level as well as by small molecule modulators and covalent modification. The human genome contains the genes for two different ACCs—ACACA and ACACB.

Aspartate kinase or aspartokinase (AK) is an enzyme that catalyzes the phosphorylation of the amino acid aspartate. This reaction is the first step in the biosynthesis of three other amino acids: methionine, lysine, and threonine, known as the "aspartate family". Aspartokinases are present only in microorganisms and plants, but not in animals, which must obtain aspartate-family amino acids from their diet. Consequently, methionine, lysine and threonine are essential amino acids in animals.
Lipoyl synthase is an enzyme that belongs to the radical SAM (S-adenosyl methionine) family. Within the radical SAM superfamily, lipoyl synthase is in a sub-family of enzymes that catalyze sulfur insertion reactions. Enzymes in this family contain two 4Fe-4S clusters, from which they obtain the sulfur groups that will be transferred onto the corresponding substrates. This particular enzyme participates in lipoic acid metabolism, so it transfers two sulfur atoms from its 4Fe-4S cluster onto the protein N6-(octanoyl)lysine through radical generation. This enzyme is usually localized to the mitochondria. Two organisms that have been extensively studied with regards to this enzyme are Escherichia coli and Mycobacterium tuberculosis. It is also found in other organisms, such as yeast and plants.

The enzyme diaminopimelate decarboxylase (EC 4.1.1.20) catalyzes the cleavage of carbon-carbon bonds in meso 2,6 diaminoheptanedioate to produce CO2 and L-lysine, the essential amino acid. It employs the cofactor pyridoxal phosphate, also known as PLP, which participates in numerous enzymatic transamination, decarboxylation and deamination reactions.
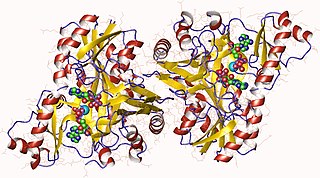
In enzymology, a biotin carboxylase (EC 6.3.4.14) is an enzyme that catalyzes the chemical reaction
In enzymology, a long-chain-fatty-acid—[acyl-carrier-protein] ligase is an enzyme that catalyzes the chemical reaction
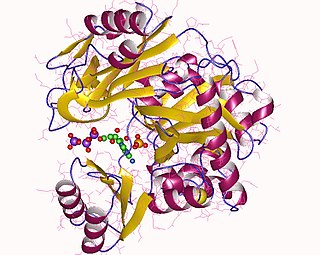
Phosphoribosylamine—glycine ligase, also known as glycinamide ribonucleotide synthetase (GARS), (EC 6.3.4.13) is an enzyme that catalyzes the chemical reaction
In enzymology, a [acyl-carrier-protein] S-malonyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a beta-ketoacyl-acyl-carrier-protein synthase I is an enzyme that catalyzes the chemical reaction
In enzymology, a lipoyl(octanoyl) transferase (EC 2.3.1.181) is an enzyme that catalyzes the chemical reaction

In enzymology and molecular biology, a holo-[acyl-carrier-protein] synthase is an enzyme that catalyzes the chemical reaction:
Biotin/lipoyl attachment domain has a conserved lysine residue that binds biotin or lipoic acid. Biotin plays a catalytic role in some carboxyl transfer reactions and is covalently attached, via an amide bond, to a lysine residue in enzymes requiring this coenzyme. Lipoamide acyltransferases have an essential cofactor, lipoic acid, which is covalently bound via an amide linkage to a lysine group. The lipoic acid cofactor is found in a variety of proteins.
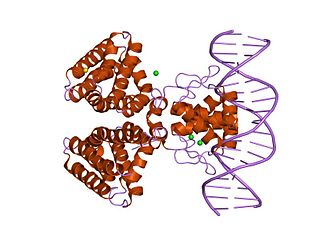
In molecular biology, the fatty acid metabolism regulator protein FadR, is a bacterial transcription factor.
Radical SAM is a designation for a superfamily of enzymes that use a [4Fe-4S]+ cluster to reductively cleave S-adenosyl-L-methionine (SAM) to generate a radical, usually a 5′-deoxyadenosyl radical (5'-dAdo), as a critical intermediate. These enzymes utilize this radical intermediate to perform diverse transformations, often to functionalize unactivated C-H bonds. Radical SAM enzymes are involved in cofactor biosynthesis, enzyme activation, peptide modification, post-transcriptional and post-translational modifications, metalloprotein cluster formation, tRNA modification, lipid metabolism, biosynthesis of antibiotics and natural products etc. The vast majority of known radical SAM enzymes belong to the radical SAM superfamily, and have a cysteine-rich motif that matches or resembles CxxxCxxC. rSAMs comprise the largest superfamily of metal-containing enzymes.
Lipoyl amidotransferase (EC 2.3.1.200, LipL (gene)) is an enzyme with systematic name (glycine cleavage system H)-N6-lipoyl-L-lysine:(lipoyl-carrier protein)-N6-L-lysine lipoyltransferase. This enzyme catalyses the following chemical reaction
Octanoyl-(GcvH):protein N-octanoyltransferase (EC 2.3.1.204, LIPL, octanoyl-[GcvH]:E2 amidotransferase, YWFL (gene)) is an enzyme with systematic name (glycine cleavage system H)-N6-octanoyl-L-lysine:(lipoyl-carrier protein)-N6-L-lysine octanoyltransferase. This enzyme catalyses the following chemical reaction
Lipoate–protein ligase (EC 2.7.7.63, LplA, lipoate protein ligase, lipoate–protein ligase A, LPL, LPL-B) is an enzyme with systematic name ATP:lipoate adenylyltransferase. This enzyme catalyses the following chemical reaction
The enzyme Pimelyl-[acyl-carrier protein] methyl ester esterase (EC 3.1.1.85, BioH; systematic name pimelyl-[acyl-carrier protein] methyl ester hydrolase catalyses the reaction
3-hydroxydecanoyl-(acyl-carrier-protein) dehydratase (EC 4.2.1.60, D-3-hydroxydecanoyl-[acyl-carrier protein] dehydratase, 3-hydroxydecanoyl-acyl carrier protein dehydrase, 3-hydroxydecanoyl-acyl carrier protein dehydratase, β-hydroxydecanoyl thioester dehydrase, β-hydroxydecanoate dehydrase, beta-hydroxydecanoyl thiol ester dehydrase, FabA, β-hydroxyacyl-acyl carrier protein dehydratase, HDDase, β-hydroxyacyl-ACP dehydrase, (3R)-3-hydroxydecanoyl-[acyl-carrier-protein] hydro-lyase) is an enzyme with systematic name (3R)-3-hydroxydecanoyl-(acyl-carrier protein) hydro-lyase. This enzyme catalyses the following chemical reaction









