
Histidine (symbol His or H) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated –NH3+ form under biological conditions), a carboxylic acid group (which is in the deprotonated –COO− form under biological conditions), and an imidazole side chain (which is partially protonated), classifying it as a positively charged amino acid at physiological pH. Initially thought essential only for infants, it has now been shown in longer-term studies to be essential for adults also. It is encoded by the codons CAU and CAC.
A condensation reaction is a class of organic addition reaction that typically proceeds in a step-wise fashion to produce the addition product, usually in equilibrium, and a water molecule. The reaction may otherwise involve the functional groups of the molecule, and formation of a small molecule such as ammonia, ethanol, or acetic acid instead of water. It is a versatile class of reactions that can occur in acidic or basic conditions or in the presence of a catalyst. This class of reactions is a vital part of life as it is essential to the formation of peptide bonds between amino acids and the biosynthesis of fatty acids.

Alamethicin is a channel-forming peptide antibiotic, produced by the fungus Trichoderma viride. It belongs to peptaibol peptides which contain the non-proteinogenic amino acid residue Aib. This residue strongly induces formation of alpha-helical structure. The peptide sequence is:

Teicoplanin is an antibiotic used in the prophylaxis and treatment of serious infections caused by Gram-positive bacteria, including methicillin-resistant Staphylococcus aureus and Enterococcus faecalis. It is a semisynthetic glycopeptide antibiotic with a spectrum of activity similar to vancomycin. Its mechanism of action is to inhibit bacterial cell wall synthesis.
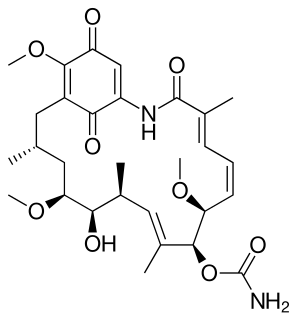
Polyketides are a large group of secondary metabolites which either contain alternating carbonyl groups and methylene groups (-CO-CH2-), or are derived from precursors which contain such alternating groups. Many polyketides are medicinal or exhibit acute toxicity.

DD-transpeptidase is a bacterial enzyme that catalyzes the transfer of the R-L-aca-D-alanyl moiety of R-L-aca-D-alanyl-D-alanine carbonyl donors to the γ-OH of their active-site serine and from this to a final acceptor. It is involved in bacterial cell wall biosynthesis, namely, the transpeptidation that crosslinks the peptide side chains of peptidoglycan strands.
Nonribosomal peptides (NRP) are a class of peptide secondary metabolites, usually produced by microorganisms like bacteria and fungi. Nonribosomal peptides are also found in higher organisms, such as nudibranchs, but are thought to be made by bacteria inside these organisms. While there exist a wide range of peptides that are not synthesized by ribosomes, the term nonribosomal peptide typically refers to a very specific set of these as discussed in this article.
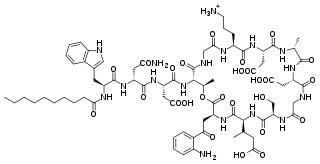
Daptomycin, sold under the brand name Cubicin among others, is a lipopeptide antibiotic used in the treatment of systemic and life-threatening infections caused by Gram-positive organisms. It is a naturally occurring compound found in the soil saprotroph Streptomyces roseosporus. Its distinct mechanism of action makes it useful in treating infections caused by multiple drug-resistant bacteria.

Viomycin is a member of the tuberactinomycin family, a group of nonribosomal peptide antibiotics exhibiting anti-tuberculosis activity. The tuberactinomycin family is an essential component in the drug cocktail currently used to fight infections of Mycobacterium tuberculosis. Viomycin was the first member of the tuberactinomycins to be isolated and identified, and was used to treat TB until it was replaced by the less toxic, but structurally related compound, capreomycin. The tuberactinomycins target bacterial ribosomes, binding RNA and disrupting bacterial protein synthesis and certain forms of RNA splicing. Viomycin is produced by the actinomycete Streptomyces puniceus.
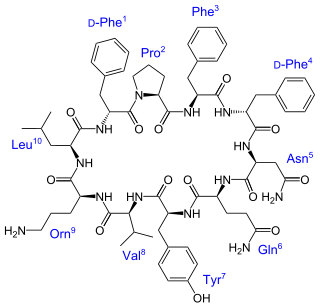
Tyrocidine is a mixture of cyclic decapeptides produced by the bacteria Bacillus brevis found in soil. It can be composed of 4 different amino acid sequences, giving tyrocidine A–D. Tyrocidine is the major constituent of tyrothricin, which also contains gramicidin. Tyrocidine was the first commercially available antibiotic, but has been found to be toxic toward human blood and reproductive cells. The function of tyrocidine within its host B. brevis is thought to be regulation of sporulation.

EF-Tu is a prokaryotic elongation factor responsible for catalyzing the binding of an aminoacyl-tRNA (aa-tRNA) to the ribosome. It is a G-protein, and facilitates the selection and binding of an aa-tRNA to the A-site of the ribosome. As a reflection of its crucial role in translation, EF-Tu is one of the most abundant and highly conserved proteins in prokaryotes. It is found in eukaryotic mitochrondria as TUFM.
Streptogramin A is a group of antibiotics within the larger family of antibiotics known as streptogramins. They are synthesized by the bacteria Streptomyces virginiae. The streptogramin family of antibiotics consists of two distinct groups: group A antibiotics contain a 23-membered unsaturated ring with lactone and peptide bonds while group B antibiotics are depsipeptides. While structurally different, these two groups of antibiotics act synergistically, providing greater antibiotic activity than the combined activity of the separate components. These antibiotics have until recently been commercially manufactured as feed additives in agriculture, although today there is increased interest in their ability to combat antibiotic-resistant bacteria, particularly vancomycin-resistant bacteria.
Streptogramin B is a subgroup of the streptogramin antibiotics family. These natural products are cyclic hexa- or hepta depsipeptides produced by various members of the genus of bacteria Streptomyces. Many of the members of the streptogramins reported in the literature have the same structure and different names; for example, pristinamycin IA = vernamycin Bα = mikamycin B = osteogrycin B.
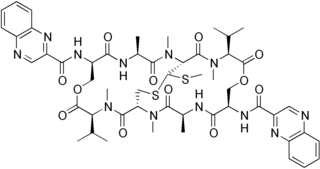
Echinomycin is a peptide antibiotic. It intercalates into DNA at two specific sites, thereby blocking the binding of hypoxia inducible factor 1 alpha (HIF1alpha).
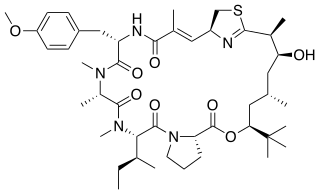
Apratoxin A - is a cyanobacterial secondary metabolite, known as a potent cytotoxic marine natural product. It is a derivative of the Apratoxin family of cytotoxins. The mixed peptide-polyketide natural product comes from a polyketide synthase/non-ribosomal peptide synthase pathway (PKS/NRPS). This cytotoxin is known for inducing G1-phase cell cycle arrest and apoptosis. This natural product's activity has made it a popular target for developing anticancer derivatives.
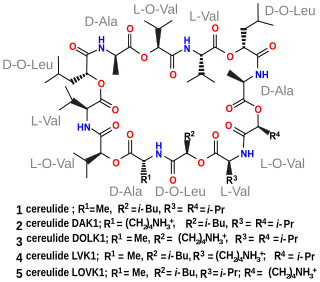
Cereulide is a toxin produced by some strains of Bacillus cereus. It is a potent cytotoxin that destroys mitochondria. It also causes nausea and vomiting.
The nonribosomal code refers to key amino acid residues and their positions within the primary sequence of an adenylation domain of a nonribosomal peptide synthetase used to predict substrate specificity and thus (partially) the final product. Analogous to the nonribosomal code is prediction of peptide composition by DNA/RNA codon reading, which is well supported by the central dogma of molecular biology and accomplished using the genetic code simply by following the DNA codon table or RNA codon table. However, prediction of natural product/secondary metabolites by the nonribosomal code is not as concrete as DNA/RNA codon-to-amino acid and much research is still needed to have a broad-use code. The increasing number of sequenced genomes and high-throughput prediction software has allowed for better elucidation of predicted substrate specificity and thus natural products/secondary metabolites. Enzyme characterization by, for example, ATP-pyrophosphate exchange assays for substrate specificity, in silico substrate-binding pocket modelling and structure-function mutagenesis helps support predictive algorithms. Much research has been done on bacteria and fungi, with prokaryotic bacteria having easier-to-predict products.
Ribosomally synthesized and post-translationally modified peptides (RiPPs), also known as ribosomal natural products, are a diverse class of natural products of ribosomal origin. Consisting of more than 20 sub-classes, RiPPs are produced by a variety of organisms, including prokaryotes, eukaryotes, and archaea, and they possess a wide range of biological functions.

The xenortides (A-D) are a class of linear peptides isolated from the bacterium Xenorhabdus nematophila, a symbiont of the entomopathogenic nematode Steinernema carpocapsae. This class of compounds is known for their insect virulence and cytotoxic biological activities. The tryptamide containing compounds show higher biological activity than the phenylethylamides. The most biologically active compound was found to be xenortide B with a potency of less than 1.6 μM activity against Trypanosoma brucei rhodesiense and Plasmodium falciparum (malaria), however it is also the most toxic to mammalian cells which limits its viability as a treatment.

Vibriobactin is a catechol siderophore that helps the microbial system to acquire iron. It was first isolated from Vibrio cholerae.














