
In biochemistry, the DNA methyltransferase family of enzymes catalyze the transfer of a methyl group to DNA. DNA methylation serves a wide variety of biological functions. All the known DNA methyltransferases use S-adenosyl methionine (SAM) as the methyl donor.

A protein family is a group of evolutionarily-related proteins. In many cases a protein family has a corresponding gene family, in which each gene encodes a corresponding protein with a 1:1 relationship. The term protein family should not be confused with family as it is used in taxonomy.
In biology and biochemistry, protease inhibitors, or antiproteases, are molecules that inhibit the function of proteases. Many naturally occurring protease inhibitors are proteins.

The Rossmann fold is a tertiary fold found in proteins that bind nucleotides, such as enzyme cofactors FAD, NAD+, and NADP+. This fold is composed of alternating beta strands and alpha helical segments where the beta strands are hydrogen bonded to each other forming an extended beta sheet and the alpha helices surround both faces of the sheet to produce a three-layered sandwich. The classical Rossmann fold contains six beta strands whereas Rossmann-like folds, sometimes referred to as Rossmannoid folds, contain only five strands. The initial beta-alpha-beta (bab) fold is the most conserved segment of the Rossmann fold. The motif is named after Michael Rossmann who first noticed this structural motif in the enzyme lactate dehydrogenase in 1970 and who later observed that this was a frequently occurring motif in nucleotide binding proteins.
Sialyltransferases are enzymes that transfer sialic acid to nascent oligosaccharide. Each sialyltransferase is specific for a particular sugar substrate. Sialyltransferases add sialic acid to the terminal portions of the sialylated glycolipids (gangliosides) or to the N- or O-linked sugar chains of glycoproteins.

Dopamine beta-hydroxylase (DBH), also known as dopamine beta-monooxygenase, is an enzyme that in humans is encoded by the DBH gene. Dopamine beta-hydroxylase catalyzes the conversion of dopamine to norepinephrine.
In enzymology, a peptidylglycine monooxygenase (EC 1.14.17.3) is an enzyme that catalyzes the chemical reaction

Procollagen-proline dioxygenase, commonly known as prolyl hydroxylase, is a member of the class of enzymes known as alpha-ketoglutarate-dependent hydroxylases. These enzymes catalyze the incorporation of oxygen into organic substrates through a mechanism that requires alpha-Ketoglutaric acid, Fe2+, and ascorbate. This particular enzyme catalyzes the formation of (2S, 4R)-4-hydroxyproline, a compound that represents the most prevalent post-translational modification in the human proteome.
In enzymology, a peptidylamidoglycolate lyase is an enzyme that catalyzes the chemical reaction

Peptidyl-glycine alpha-amidating monooxygenase is an enzyme that catalyzes the conversion of glycine amides to amides and glyoxylate.
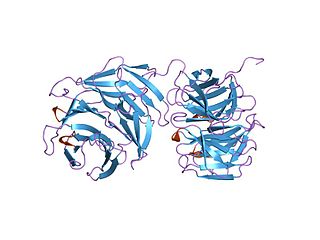
The NHL repeat, named after ncl-1, HT2A and lin-41, is an amino acid sequence found largely in a large number of eukaryotic and prokaryotic proteins. For example, the repeat is found in a variety of enzymes of the copper type II, ascorbate-dependent monooxygenase family which catalyse the C-terminus alpha-amidation of biological peptides. In many it occurs in tandem arrays, for example in the RING finger beta-box, coiled-coil (RBCC) eukaryotic growth regulators. The arthropod 'Brain Tumor' protein is one such growth regulator that contains a 6-bladed NHL-repeat beta-propeller.
The aminoacyl-tRNA synthetases catalyse the attachment of an amino acid to its cognate transfer RNA molecule in a highly specific two-step reaction. These proteins differ widely in size and oligomeric state, and have limited sequence homology. The 20 aminoacyl-tRNA synthetases are divided into two classes, I and II. Class I aminoacyl-tRNA synthetases contain a characteristic Rossmann fold catalytic domain and are mostly monomeric. Class II aminoacyl-tRNA synthetases share an anti-parallel beta-sheet fold flanked by alpha-helices, and are mostly dimeric or multimeric, containing at least three conserved regions. However, tRNA binding involves an alpha-helical structure that is conserved between class I and class II synthetases. In reactions catalysed by the class I aminoacyl-tRNA synthetases, the aminoacyl group is coupled to the 2'-hydroxyl of the tRNA, while, in class II reactions, the 3'-hydroxyl site is preferred. The synthetases specific for arginine, cysteine, glutamic acid, glutamine, isoleucine, leucine, methionine, tyrosine, tryptophan and valine belong to class I synthetases; these synthetases are further divided into three subclasses, a, b and c, according to sequence homology. The synthetases specific for alanine, asparagine, aspartic acid, glycine, histidine, lysine, phenylalanine, proline, serine, and threonine belong to class-II synthetases.

DBH-like monooxygenase protein 1, also known as monooxygenase X, is an enzyme that in humans is encoded by the MOXD1 gene.

S-adenosylmethionine synthetase is an enzyme that creates S-adenosylmethionine by reacting methionine and ATP.
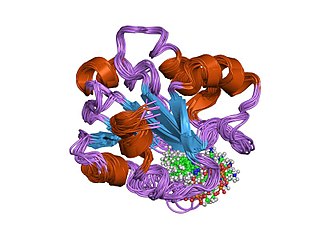
In molecular biology, the vitamin B12-binding domain is a protein domain which binds to cobalamin. It can bind two different forms of the cobalamin cofactor, with cobalt bonded either to a methyl group (methylcobalamin) or to 5'-deoxyadenosine (adenosylcobalamin). Cobalamin-binding domains are mainly found in two families of enzymes present in animals and prokaryotes, which perform distinct kinds of reactions at the cobalt-carbon bond. Enzymes that require methylcobalamin carry out methyl transfer reactions. Enzymes that require adenosylcobalamin catalyse reactions in which the first step is the cleavage of adenosylcobalamin to form cob(II)alamin and the 5'-deoxyadenosyl radical, and thus act as radical generators. In both types of enzymes the B12-binding domain uses a histidine to bind the cobalt atom of cobalamin cofactors. This histidine is embedded in a DXHXXG sequence, the most conserved primary sequence motif of the domain. Proteins containing the cobalamin-binding domain include:
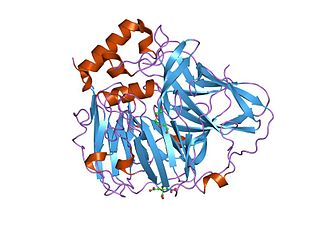
In molecular biology, multicopper oxidases are enzymes which oxidise their substrate by accepting electrons at a mononuclear copper centre and transferring them to a trinuclear copper centre; dioxygen binds to the trinuclear centre and, following the transfer of four electrons, is reduced to two molecules of water. There are three spectroscopically different copper centres found in multicopper oxidases: type 1, type 2 and type 3. Multicopper oxidases consist of 2, 3 or 6 of these homologous domains, which also share homology with the cupredoxins azurin and plastocyanin. Structurally, these domains consist of a cupredoxin-like fold, a beta-sandwich consisting of 7 strands in 2 beta-sheets, arranged in a Greek-key beta-barrel. Multicopper oxidases include:

In molecular biology, Glycoside hydrolase family 14 is a family of glycoside hydrolases.
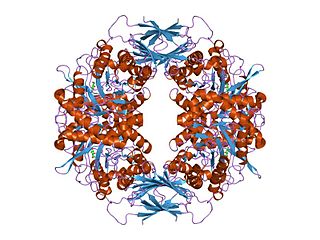
In molecular biology, glycoside hydrolase family 39 is a family of glycoside hydrolases.
A protein superfamily is the largest grouping (clade) of proteins for which common ancestry can be inferred. Usually this common ancestry is inferred from structural alignment and mechanistic similarity, even if no sequence similarity is evident. Sequence homology can then be deduced even if not apparent. Superfamilies typically contain several protein families which show sequence similarity within each family. The term protein clan is commonly used for protease and glycosyl hydrolases superfamilies based on the MEROPS and CAZy classification systems.

The flavin-containing monooxygenase (FMO) protein family specializes in the oxidation of xeno-substrates in order to facilitate the excretion of these compounds from living organisms. These enzymes can oxidize a wide array of heteroatoms, particularly soft nucleophiles, such as amines, sulfides, and phosphites. This reaction requires an oxygen, an NADPH cofactor, and an FAD prosthetic group. FMOs share several structural features, such as a NADPH binding domain, FAD binding domain, and a conserved arginine residue present in the active site. Recently, FMO enzymes have received a great deal of attention from the pharmaceutical industry both as a drug target for various diseases and as a means to metabolize pro-drug compounds into active pharmaceuticals. These monooxygenases are often misclassified because they share activity profiles similar to those of cytochrome P450 (CYP450), which is the major contributor to oxidative xenobiotic metabolism. However, a key difference between the two enzymes lies in how they proceed to oxidize their respective substrates; CYP enzymes make use of an oxygenated heme prosthetic group, while the FMO family utilizes FAD to oxidize its substrates.















