Related Research Articles

In genetics, complementary DNA (cDNA) is DNA synthesized from a single-stranded RNA template in a reaction catalyzed by the enzyme reverse transcriptase. cDNA is often used to clone eukaryotic genes in prokaryotes. When scientists want to express a specific protein in a cell that does not normally express that protein, they will transfer the cDNA that codes for the protein to the recipient cell. cDNA is also produced naturally by retroviruses and then integrated into the host's genome, where it creates a provirus.

A primer is a short single-stranded nucleic acid utilized by all living organisms in the initiation of DNA synthesis. The enzymes responsible for DNA replication, DNA polymerases, are only capable of adding nucleotides to the 3’-end of an existing nucleic acid, requiring a primer be bound to the template before DNA polymerase can begin a complementary strand. Living organisms use solely RNA primers, while laboratory techniques in biochemistry and molecular biology that require in vitro DNA synthesis usually use DNA primers, since they are more temperature stable.

Protein biosynthesis is a core biological process, occurring inside cells, balancing the loss of cellular proteins through the production of new proteins. Proteins perform a variety of critical functions as enzymes, structural proteins or hormones and therefore, are crucial biological components. Protein synthesis is a very similar process for both prokaryotes and eukaryotes but there are some distinct differences.

A retrovirus is a type of RNA virus that inserts a copy of its genome into the DNA of a host cell that it invades, thus changing the genome of that cell. Once inside the host cell's cytoplasm, the virus uses its own reverse transcriptase enzyme to produce DNA from its RNA genome, the reverse of the usual pattern, thus retro (backwards). The new DNA is then incorporated into the host cell genome by an integrase enzyme, at which point the retroviral DNA is referred to as a provirus. This integration of genome to the host cell allows retroviruses to have many unique features such as ability to cause cancer, permanently change the host genome and ability to introduce desired transgenes as permanently within gene therapy. The host cell then treats the viral DNA as part of its own genome, transcribing and translating the viral genes along with the cell's own genes, producing the proteins required to assemble new copies of the virus.

A reverse transcriptase (RT) is an enzyme used to generate complementary DNA (cDNA) from an RNA template, a process termed reverse transcription. Reverse transcriptases are used by retroviruses to replicate their genomes, by retrotransposon mobile genetic elements to proliferate within the host genome, by eukaryotic cells to extend the telomeres at the ends of their linear chromosomes, and by some non-retroviruses such as the hepatitis B virus, a member of the Hepadnaviridae, which are dsDNA-RT viruses.

Transcription is the first of several steps of DNA based gene expression in which a particular segment of DNA is copied into RNA by the enzyme RNA polymerase.

RNA polymerase, abbreviated RNAP or RNApol, officially DNA-directed RNA polymerase, is an enzyme that synthesizes RNA from a DNA template. RNAP locally opens the double-stranded DNA so that one strand of the exposed nucleotides can be used as a template for the synthesis of RNA, a process called transcription. A transcription factor and its associated transcription mediator complex must be attached to a DNA binding site called a promoter region before RNAP can initiate the DNA unwinding at that position. RNAP not only initiates RNA transcription, it also guides the nucleotides into position, facilitates attachment and elongation, has intrinsic proofreading and replacement capabilities, and termination recognition capability. In eukaryotes, RNAP can build chains as long as 2.4 million nucleotides.
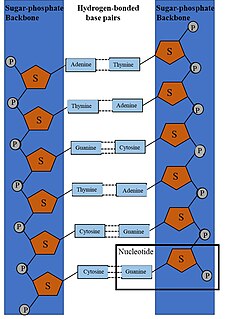
DNA synthesis is the natural or artificial creation of deoxyribonucleic acid (DNA) molecules. DNA is a macromolecule made up of nucleotide units, which are linked by covalent bonds and hydrogen bonds, in a repeating structure. DNA synthesis occurs when these nucelotide units are joined together to form DNA; this can occur artificially or naturally. Nucleotide units are made up of a nitrogenous base, pentose sugar (deoxyribose) and phosphate group. Each unit is joined when a covalent bond forms between its phosphate group and the pentose sugar of the next nucleotide, forming a sugar-phosphate backbone. DNA is a complementary, double stranded structure as specific base pairing occurs naturally when hydrogen bonds form between the nucleotide bases.

A ρ factor is a prokaryotic protein involved in the termination of transcription. Rho factor binds to the transcription terminator pause site, an exposed region of single stranded RNA after the open reading frame at C-rich/G-poor sequences that lack obvious secondary structure.
In genetics, a transcription terminator is a section of nucleic acid sequence that marks the end of a gene or operon in genomic DNA during transcription. This sequence mediates transcriptional termination by providing signals in the newly synthesized transcript RNA that trigger processes which release the transcript RNA from the transcriptional complex. These processes include the direct interaction of the mRNA secondary structure with the complex and/or the indirect activities of recruited termination factors. Release of the transcriptional complex frees RNA polymerase and related transcriptional machinery to begin transcription of new mRNAs.
The Pribnow box is the sequence TATAAT of six nucleotides that is an essential part of a promoter site on DNA for transcription to occur in bacteria. It is an idealized or consensus sequence—that is, it shows the most frequently occurring base at each position in many promoters analyzed; individual promoters often vary from the consensus at one or more positions. It is also commonly called the -10 sequence, because it is centered roughly ten base pairs upstream from the site of initiation of transcription.

Retrotransposons are a type of genetic component that copy and paste themselves into different genomic locations (transposon) by converting RNA back into DNA through the process reverse transcription using an RNA transposition intermediate.
This is a list of topics in molecular biology. See also index of biochemistry articles.

In molecular genetics, a repressor is a DNA- or RNA-binding protein that inhibits the expression of one or more genes by binding to the operator or associated silencers. A DNA-binding repressor blocks the attachment of RNA polymerase to the promoter, thus preventing transcription of the genes into messenger RNA. An RNA-binding repressor binds to the mRNA and prevents translation of the mRNA into protein. This blocking of expression is called repression.
Attenuation is a proposed mechanism of control in some bacterial operons which results in premature termination of transcription and is based on the fact that, in bacteria, transcription and translation proceed simultaneously. Attenuation involves a provisional stop signal (attenuator), located in the DNA segment that corresponds to the leader sequence of mRNA. During attenuation, the ribosome becomes stalled (delayed) in the attenuator region in the mRNA leader. Depending on the metabolic conditions, the attenuator either stops transcription at that point or allows read-through to the structural gene part of the mRNA and synthesis of the appropriate protein.
Antitermination is the prokaryotic cell's aid to fix premature termination of RNA synthesis during the transcription of RNA. It occurs when the RNA polymerase ignores the termination signal and continues elongating its transcript until a second signal is reached. Antitermination provides a mechanism whereby one or more genes at the end of an operon can be switched either on or off, depending on the polymerase either recognizing or not recognizing the termination signal.
This glossary of genetics is a list of definitions of terms and concepts commonly used in the study of genetics and related disciplines in biology, including molecular biology and evolutionary biology. It is intended as introductory material for novices; for more specific and technical detail, see the article corresponding to each term. For related terms, see Glossary of evolutionary biology.

Bacterial transcription is the process in which a segment of bacterial DNA is copied into a newly synthesized strand of messenger RNA (mRNA) with use of the enzyme RNA polymerase. The process occurs in three main steps: initiation, elongation, and termination; and the end result is a strand of mRNA that is complementary to a single strand of DNA. Generally, the transcribed region accounts for more than one gene. In fact, many prokaryotic genes occur in operons, which are a series of genes that work together to code for the same protein or gene product and are controlled by a single promoter. Bacterial RNA polymerase is made up of four subunits and when a fifth subunit attaches, called the σ-factor, the polymerase can recognize specific binding sequences in the DNA, called promoters. The binding of the σ-factor to the promoter is the first step in initiation. Once the σ-factor releases from the polymerase, elongation proceeds. The polymerase continues down the double stranded DNA, unwinding it and synthesizing the new mRNA strand until it reaches a termination site. There are two termination mechanisms that are discussed in further detail below. Termination is required at specific sites for proper gene expression to occur. Gene expression determines how much gene product, such as protein, is made by the gene. Transcription is carried out by RNA polymerase but its specificity is controlled by sequence-specific DNA binding proteins called transcription factors. Transcription factors work to recognize specific DNA sequences and based on the cells needs, promote or inhibit additional transcription.

Rho-independent termination is a mechanism in prokaryotes that causes RNA transcription to stop and release the newly made RNA. In this mechanism, the mRNA contains a sequence that can base pair with itself to form a stem-loop structure 7–20 base pairs in length that is also rich in cytosine-guanine base pairs. C-G base pairs have significant base-stacking interactions and can form three hydrogen bonds between each other, resulting in a stable RNA duplex. Following the stem-loop structure is a chain of uracil residues. The bonds between uracil and adenine are very weak. A protein bound to RNA polymerase (nusA) binds to the stem-loop structure tightly enough to cause the polymerase to temporarily stall. This pausing of the polymerase coincides with transcription of the poly-uracil sequence. The weak adenine-uracil bonds lower the energy of destabilization for the RNA-DNA duplex, allowing it to unwind and dissociate from the RNA polymerase.
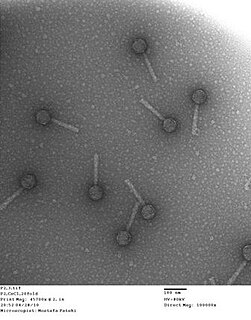
Bacteriophage P2, scientific name Escherichia virus P2, is a temperate phage that infects E. coli. It is a tailed virus with a contractile sheath and is thus classified in the genus Peduovirus, subfamily Peduovirinae, family Myoviridae within order Caudovirales. This genus of viruses includes many P2-like phages as well as the satellite phage P4.
References
- ↑ Richard Calendar (2006) The Bacteriophages (Oxford University Press, Oxford, NY) ISBN 0-19-514850-9, page 67