The term allele denotes the variant of a given gene. In genetics it is normal for genes to show deviations or diversity − all alleles together make up the set of genetic information that defines a gene. For example, the ABO blood grouping is controlled by the ABO gene, which has six common variants (alleles). In population genetics nearly every living human's phenotype for the ABO gene is some combination of just these six alleles.
A microsatellite is a tract of repetitive DNA in which certain DNA motifs are repeated, typically 5–50 times. Microsatellites occur at thousands of locations within an organism's genome. They have a higher mutation rate than other areas of DNA leading to high genetic diversity. Microsatellites are often referred to as short tandem repeats (STRs) by forensic geneticists and in genetic genealogy, or as simple sequence repeats (SSRs) by plant geneticists.
Allele frequency, or gene frequency, is the relative frequency of an allele at a particular locus in a population, expressed as a fraction or percentage. Specifically, it is the fraction of all chromosomes in the population that carry that allele. Microevolution is the change in allele frequencies that occurs over time within a population.

In genetics, a single-nucleotide polymorphism is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in a sufficiently large fraction of the population, many publications do not apply such a frequency threshold.

A haplotype is a group of alleles in an organism that are inherited together from a single parent.
A quantitative trait locus (QTL) is a locus that correlates with variation of a quantitative trait in the phenotype of a population of organisms. QTLs are mapped by identifying which molecular markers correlate with an observed trait. This is often an early step in identifying and sequencing the actual genes that cause the trait variation.
Human Genetic Diversity: Lewontin's Fallacy is a 2003 paper by A. W. F. Edwards. He criticises an argument first made in Richard Lewontin's 1972 article "The Apportionment of Human Diversity", that the practice of dividing humanity into races is taxonomically invalid because any given individual will often have more in common genetically with members of other population groups than with members of their own. Edwards argued that this does not refute the biological reality of race since genetic analysis can usually make correct inferences about the perceived race of a person from whom a sample is taken, and that the rate of success increases when more genetic loci are examined.
A Y-STR is a short tandem repeat (STR) on the Y-chromosome. Y-STRs are often used in forensics, paternity, and genealogical DNA testing. Y-STRs are taken specifically from the male Y chromosome. These Y-STRs provide a weaker analysis than autosomal STRs because the Y chromosome is only found in males, which are only passed down by the father, making the Y chromosome in any paternal line practically identical. This causes a significantly smaller amount of distinction between Y-STR samples. Autosomal STRs provide a much stronger analytical power because of the random matching that occurs between pairs of chromosomes during the zygote making process.
Genetic association is when one or more genotypes within a population co-occur with a phenotypic trait more often than would be expected by chance occurrence.
Second Generation Multiplex Plus , is a DNA profiling system developed by Applied Biosystems. It is an updated version of Second Generation Multiplex. SGM Plus has been used by the UK National DNA Database since 1998.
Short Tandem Repeat (STR) analysis is a common molecular biology method used to compare allele repeats at specific loci in DNA between two or more samples. A short tandem repeat is a microsatellite with repeat units that are 2 to 7 base pairs in length, with the number of repeats varying among individuals, making STRs effective for human identification purposes. This method differs from restriction fragment length polymorphism analysis (RFLP) since STR analysis does not cut the DNA with restriction enzymes. Instead, polymerase chain reaction (PCR) is employed to discover the lengths of the short tandem repeats based on the length of the PCR product.

In population genetics, the Balding–Nichols model is a statistical description of the allele frequencies in the components of a sub-divided population. With background allele frequency p the allele frequencies, in sub-populations separated by Wright's FSTF, are distributed according to independent draws from
A DNA database or DNA databank is a database of DNA profiles which can be used in the analysis of genetic diseases, genetic fingerprinting for criminology, or genetic genealogy. DNA databases may be public or private, the largest ones being national DNA databases.
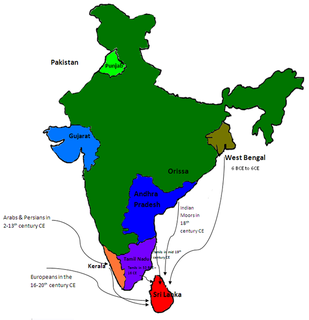
Genetic studies on the Sinhalese is part of population genetics investigating the origins of the Sinhalese population.
In paternity testing, Paternity Index (PI) is a calculated value generated for a single genetic marker or locus and is associated with the statistical strength or weight of that locus in favor of or against parentage given the phenotypes of the tested participants and the inheritance scenario. Phenotype typically refers to physical characteristics such as body plan, color, behavior, etc. in organisms. However, the term used in the area of DNA paternity testing refers to what is observed directly in the laboratory. Laboratories involved in parentage testing and other fields of human identity employ genetic testing panels that contain a battery of loci each of which is selected due to extensive allelic variations within and between populations. These genetic variations are not assumed to bestow physical and/or behavioral attributes to the person carrying the allelic arrangement(s) and therefore are not subject to selective pressure and follow Hardy Weinberg inheritance patterns.

The Combined DNA Index System (CODIS) is the United States national DNA database created and maintained by the Federal Bureau of Investigation. CODIS consists of three levels of information; Local DNA Index Systems (LDIS) where DNA profiles originate, State DNA Index Systems (SDIS) which allows for laboratories within states to share information, and the National DNA Index System (NDIS) which allows states to compare DNA information with one another.
Although Sri Lankan Tamils are culturally and linguistically distinct, genetic studies indicate that they are closely related to other ethnic groups in the island while being related to the Indian Tamils from South India and Bengalis from the East India as well. There are various studies that indicate varying degrees of connections between Sri Lankan Tamils, Sinhalese and Indian ethnic groups.
The relationship of the Mayas to other indigenous peoples of the Americas has been assessed using traditional genetic markers. Mayas inhabited several parts of Mexico and Central America, including Chiapas, the northern lowlands of the Yucatán Peninsula, the southern lowlands and highlands of Guatemala, Belize, and parts of western El Salvador and Honduras. Genetic studies of the Maya people are reported to show higher levels of variation when compared to other groups.

DNA profiling is the determination of a DNA profile for legal and investigative purposes. DNA analysis methods have changed numerous times over the years as technology improves and allows for more information to be determined with less starting material. Modern DNA analysis is based on the statistical calculation of the rarity of the produced profile within a population.





