
Poliovirus, the causative agent of polio, is a member virus of Enterovirus C, in the family of Picornaviridae.

A picornavirus is a virus belonging to the family Picornaviridae, a family of viruses in the order Picornavirales. Vertebrates, including humans, serve as natural hosts. Picornaviruses are nonenveloped viruses that represent a large family of small, cytoplasmic, plus-strand RNA (~7.5kb) viruses with a 30-nm icosahedral capsid. Its genome does not have a lipid membrane. Picornaviruses are found in mammals and birds. There are currently 80 species in this family, divided among 35 genera. Notable examples are Enterovirus, Aphthovirus, Cardiovirus, and Hepatovirus genera. The viruses in this family can cause a range of diseases including paralysis, meningitis, hepatitis and poliomyelitis. Picornaviruses are in Baltimore IV class. Their genome single-stranded (+) sense RNA is what functions as mRNA after entry into the cell and all viral mRNA synthesized is of genome polarity. The mRNA encodes RNA dependent RNA polymerase. This polymerase makes complementary minus strands of RNA, then uses them as templates to make more plus strands. So, an overview of the steps in picornavirus replication are in order: attachment, entry, translation, transcription/genome replication, assembly and exit.

Rubella virus (RuV) is the pathogenic agent of the disease rubella, and is the main cause of congenital rubella syndrome when infection occurs during the first weeks of pregnancy.
VPg is a protein that is covalently attached to the 5′ end of positive strand viral RNA and acts as a primer during RNA synthesis in a variety of virus families including Picornaviridae, Potyviridae and Caliciviridae. The primer activity of VPg occurs when the protein becomes uridylated, providing a free hydroxyl that can be extended by the virally encoded RNA-dependent RNA polymerase. For some virus families, VPg also has a role in translation initiation by acting like a 5' mRNA cap.

The coronavirus SL-III cis-acting replication element (CRE) is an RNA element that regulates defective interfering (DI) RNA replication.
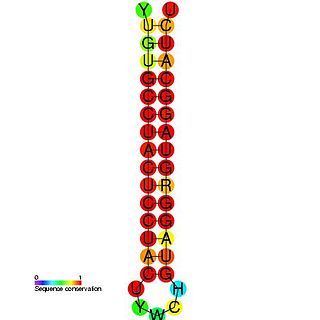
The Hepatitis C stem-loop IV is part of a putative RNA element found in the NS5B coding region. This element along with stem-loop VII, is important for colony formation, though its exact function and mechanism are unknown.

The Hepatitis C virus 3'X element is an RNA element which contains three stem-loop structures that are essential for replication.

The Hepatitis C virus (HCV) cis-acting replication element (CRE) is an RNA element which is found in the coding region of the RNA-dependent RNA polymerase NS5B. Mutations in this family have been found to cause a blockage in RNA replication and it is thought that both the primary sequence and the structure of this element are crucial for HCV RNA replication.

Retroviral Psi packaging element is a cis-acting RNA element identified in the genomes of the retroviruses Human immunodeficiency virus (HIV) and Simian immunodeficiency virus (SIV). It is involved in regulating the essential process of packaging the retroviral RNA genome into the viral capsid during replication. The final virion contains a dimer of two identical unspliced copies of the viral genome.

The Human Parechovirus 1 cis regulatory element is an RNA element which is located in the 5'-terminal 112 nucleotides of the genome of human parechovirus 1 (HPeV1). The element consists of two stem-loop structures together with a pseudoknot. Disruption of any of these elements impairs both viral replication and growth.
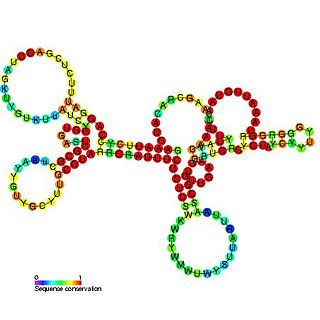
Tombusvirus 5′ UTR is an important cis-regulatory region of the Tombus virus genome.
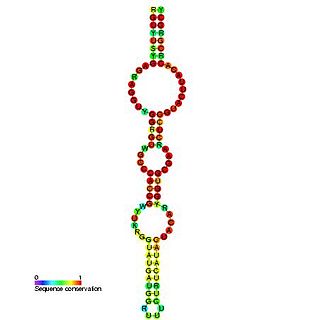
In virology, the tombusvirus internal replication element (IRE) is a segment of RNA located within the region coding for p92 polymerase. This element is essential for viral replication; specifically, it is thought to be required at an early stage of replication, such as template recruitment and/or replicase complex assembly.
Turnip crinkle virus (TCV) is a plant pathogenic virus of the family Tombusviridae. It was first isolated from turnip. TCV is a small, single-stranded, positive-sense RNA virus. It has been shown to infect various types of plant species including the common plant model, Arabidopsis thaliana. Its gRNA encodes for five proteins: p28 and p88 (replication), p8 and p9 (movement) and CP. The structure of the virus was determined to 3.2 Ångstrom resolution using x-ray crystallography in 1986. It is structurally quite similar to the tomato bushy stunt virus.

Vincent R. Racaniello is a Higgins Professor in the Department of Microbiology and Immunology at Columbia University’s College of Physicians and Surgeons. He is one of four virologists who has recently authored Principles of Virology, a textbook used by many teaching virology to undergraduate, medical and graduate students.

The Flavivirus capsid hairpin cHP is a conserved RNA hairpin structure identified within the capsid coding region of several flavivirus genomes. These positive strand RNA genomes are translated as a single polypeptide and subsequently cleaved into constituent proteins, the first of which is the capsid protein. The cHP hairpin is located within the capsid coding region between two AUG start codons. The cHP cis element has been shown to direct translation start from the suboptimal first start codon. The ability of cHP to direct initiation from the first start codon is proportional to its thermodynamic stability, is position dependent, and is sequence independent. It has been demonstrated that both AUGs and the conserved cHP are necessary for efficient viral replication in human and mosquito cells.

Poly(rC)-binding protein 1 is a protein that in humans is encoded by the PCBP1 gene.

Picornain 3C (EC 3.4.22.28, Picornain 3C is a protease and endopeptidase enzyme found in the picornavirus, that cleaves peptide bonds of non- terminal sequences. Picornain 3C’s proteinase activity is primarily responsible for the catalytic process of selectively cleaving Gln-Gly bonds in the polyprotein of poliovirus and substitution of Glu for Gln, and Ser or Thr for Gly in other picornaviruses. Picornain 3C are cysteine proteases related by amino acid sequence to trypsin-like serine proteases. Picornain 3C is encoded by enteroviruses, rhinoviruses, aphtoviruses and cardioviruses. These genera all cause a wide range of infections for humans and other mammals.
In molecular biology, the Hepatitis A virus cis-acting replication element (CRE) is an RNA element which is found in the coding region of the RNA-dependent RNA polymerase in Hepatitis A virus (HAV). It is larger than the CREs found in related Picornavirus species, but is thought to be functionally similar. It is thought to be involved in uridylylation of VPg.
In molecular biology, the Avian encephalitis virus cis-acting replication element (CRE) is an s an RNA element which is found in the coding region of the RNA-dependent RNA polymerase in Avian encephalitis virus (AEV). It is structurally similar to the Hepatitis A virus cis-acting replication element.




















