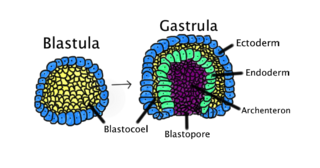
Gastrulation is the stage in the early embryonic development of most animals, during which the blastula, or in mammals the blastocyst is reorganized into a multilayered structure known as the gastrula. Before gastrulation, the embryo is a continuous epithelial sheet of cells; by the end of gastrulation, the embryo has begun differentiation to establish distinct cell lineages, set up the basic axes of the body, and internalized one or more cell types including the prospective gut.
Segmentation in biology is the division of some animal and plant body plans into a series of repetitive segments. This article focuses on the segmentation of animal body plans, specifically using the examples of the taxa Arthropoda, Chordata, and Annelida. These three groups form segments by using a "growth zone" to direct and define the segments. While all three have a generally segmented body plan and use a growth zone, they use different mechanisms for generating this patterning. Even within these groups, different organisms have different mechanisms for segmenting the body. Segmentation of the body plan is important for allowing free movement and development of certain body parts. It also allows for regeneration in specific individuals.
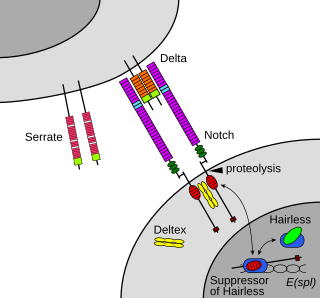
The Notch signaling pathway is a highly conserved cell signaling system present in most animals. Mammals possess four different notch receptors, referred to as NOTCH1, NOTCH2, NOTCH3, and NOTCH4. The notch receptor is a single-pass transmembrane receptor protein. It is a hetero-oligomer composed of a large extracellular portion, which associates in a calcium-dependent, non-covalent interaction with a smaller piece of the notch protein composed of a short extracellular region, a single transmembrane-pass, and a small intracellular region.

Neural crest cells are a temporary group of cells that arise from the embryonic ectoderm germ layer, and in turn give rise to a diverse cell lineage—including melanocytes, craniofacial cartilage and bone, smooth muscle, peripheral and enteric neurons and glia.

The primitive streak is a structure that forms in the early embryo in amniotes. In amphibians the equivalent structure is the blastopore. During early embryonic development, the embryonic disc becomes oval shaped, and then pear-shaped with the broad end towards the anterior, and the narrower region projected to the posterior. The primitive streak forms a longitudinal midline structure in the narrower posterior (caudal) region of the developing embryo on its dorsal side. At first formation the primitive streak extends for half the length of the embryo. In the human embryo this appears by stage 6, about 17 days.
Compartments can be simply defined as separate, different, adjacent cell populations, which upon juxtaposition, create a lineage boundary. This boundary prevents cell movement from cells from different lineages across this barrier, restricting them to their compartment. Subdivisions are established by morphogen gradients and maintained by local cell-cell interactions, providing functional units with domains of different regulatory genes, which give rise to distinct fates. Compartment boundaries are found across species. In the hindbrain of vertebrate embryos, rhombomeres are compartments of common lineage outlined by expression of Hox genes. In invertebrates, the wing imaginal disc of Drosophila provides an excellent model for the study of compartments. Although other tissues, such as the abdomen, and even other imaginal discs are compartmentalized, much of our understanding of key concepts and molecular mechanisms involved in compartment boundaries has been derived from experimentation in the wing disc of the fruit fly.
An asymmetric cell division produces two daughter cells with different cellular fates. This is in contrast to symmetric cell divisions which give rise to daughter cells of equivalent fates. Notably, stem cells divide asymmetrically to give rise to two distinct daughter cells: one copy of the original stem cell as well as a second daughter programmed to differentiate into a non-stem cell fate.
Within the field of developmental biology, one goal is to understand how a particular cell develops into a final cell type, known as fate determination. Within an embryo, several processes play out at the cellular and tissue level to create an organism. These processes include cell proliferation, differentiation, cellular movement and programmed cell death. Each cell in an embryo receives molecular signals from neighboring cells in the form of proteins, RNAs and even surface interactions. Almost all animals undergo a similar sequence of events during very early development, a conserved process known as embryogenesis. During embryogenesis, cells exist in three germ layers, and undergo gastrulation. While embryogenesis has been studied for more than a century, it was only recently that scientists discovered that a basic set of the same proteins and mRNAs are involved in embryogenesis. Evolutionary conservation is one of the reasons that model systems such as the fly, the mouse, and other organisms are used as models to study embryogenesis and developmental biology. Studying model organisms provides information relevant to other animals, including humans. While studying the different model systems, cells fate was discovered to be determined via multiple ways, two of which are by the combination of transcription factors the cells have and by the cell-cell interaction. Cells’ fate determination mechanisms were categorized into three different types, autonomously specified cells, conditionally specified cells, or syncytial specified cells. Furthermore, the cells’ fate was determined mainly using two types of experiments, cell ablation and transplantation. The results obtained from these experiments, helped in identifying the fate of the examined cells.
In the field of developmental biology, regional differentiation is the process by which different areas are identified in the development of the early embryo. The process by which the cells become specified differs between organisms.
The scleraxis protein is a member of the basic helix-loop-helix (bHLH) superfamily of transcription factors. Currently two genes have been identified to code for identical scleraxis proteins.

Fate mapping is a method used in developmental biology to study the embryonic origin of various adult tissues and structures. The "fate" of each cell or group of cells is mapped onto the embryo, showing which parts of the embryo will develop into which tissue. When carried out at single-cell resolution, this process is called cell lineage tracing. It is also used to trace the development of tumors.
Convergent extension (CE), sometimes called convergence and extension (C&E), is the process by which the tissue of an embryo is restructured to converge (narrow) along one axis and extend (elongate) along a perpendicular axis by cellular movement.

Protein numb homolog is a protein that in humans is encoded by the NUMB gene. The protein encoded by this gene plays a role in the determination of cell fates during development. The encoded protein, whose degradation is induced in a proteasome-dependent manner by MDM2, is a membrane-bound protein that has been shown to associate with EPS15, LNX1, and NOTCH1. Four transcript variants encoding different isoforms have been found for this gene.
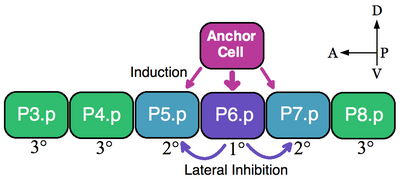
The anchor cell is a cell in nematodes such as Caenorhabditis elegans. It is important in the development of the reproductive system, as it is required for the production of the tube of cells that allows embryos to pass from the uterus through the vulva to the outside of the worm.
The trunk neural crest or truncal neural crest is one of the regions of neural crest in the embryo.
The Nodal signaling pathway is a signal transduction pathway important in regional and cellular differentiation during embryonic development.

A teloblast is a large cell in the embryos of clitellate annelids which asymmetrically divide to form many smaller cells known as blast cells. These blast cells further proliferate and differentiate to form the segmental tissues of the annelid. Teloblasts are well studied in leeches, though they are also present in the other major class of clitellates: the oligochaetes.

Retinal precursor cells are biological cells that differentiate into the various cell types of the retina during development. In the vertebrate, these retinal cells differentiate into seven cell types, including retinal ganglion cells, amacrine cells, bipolar cells, horizontal cells, rod photoreceptors, cone photoreceptors, and Müller glia cells. During embryogenesis, retinal cells originate from the anterior portion of the neural plate termed the eye field. Eye field cells with a retinal fate express several transcription factor markers including Rx1, Pax6, and Lhx2. The eye field gives rise to the optic vesicle and then to the optic cup. The retina is generated from the precursor cells within the inner layer of the optic cup, as opposed to the retinal pigment epithelium that originate from the outer layer of the optic cup. In general, the developing retina is organized so that the least-committed precursor cells are located in the periphery of the retina, while the committed cells are located in the center of the retina. The differentiation of retinal precursor cells into the mature cell types found in the retina is coordinated in time and space by factors within the cell as well as factors in the environment of the cell. One example of an intrinsic regulator of this process is the transcription factor Ath5. Ath5 expression in retinal progenitor cells biases their differentiation into a retinal ganglion cell fate. An example of an environmental factor is the morphogen sonic hedge hog (Shh). Shh has been shown to repress the differentiation of precursor cells into retinal ganglion cells.
Proneural genes encode transcription factors of the basic helix-loop-helix (bHLH) class which are responsible for the development of neuroectodermal progenitor cells. Proneural genes have multiple functions in neural development. They integrate positional information and contribute to the specification of progenitor-cell identity. From the same ectodermal cell types, neural or epidermal cells can develop based on interactions between proneural and neurogenic genes. Neurogenic genes are so called because loss of function mutants show an increase number of developed neural precursors. On the other hand, proneural genes mutants fail to develop neural precursor cells.
Paul W. Sternberg is an American biologist. He does research for WormBase on C. elegans, a model organism.