Related Research Articles

An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term exon refers to both the DNA sequence within a gene and to the corresponding sequence in RNA transcripts. In RNA splicing, introns are removed and exons are covalently joined to one another as part of generating the mature RNA. Just as the entire set of genes for a species constitutes the genome, the entire set of exons constitutes the exome.
An intron is any nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word intron is derived from the term intragenic region, i.e., a region inside a gene. The term intron refers to both the DNA sequence within a gene and the corresponding RNA sequence in RNA transcripts. The non-intron sequences that become joined by this RNA processing to form the mature RNA are called exons.

RNA splicing is a process in molecular biology where a newly-made precursor messenger RNA (pre-mRNA) transcript is transformed into a mature messenger RNA (mRNA). It works by removing all the introns and splicing back together exons. For nuclear-encoded genes, splicing occurs in the nucleus either during or immediately after transcription. For those eukaryotic genes that contain introns, splicing is usually needed to create an mRNA molecule that can be translated into protein. For many eukaryotic introns, splicing occurs in a series of reactions which are catalyzed by the spliceosome, a complex of small nuclear ribonucleoproteins (snRNPs). There exist self-splicing introns, that is, ribozymes that can catalyze their own excision from their parent RNA molecule. The process of transcription, splicing and translation is called gene expression, the central dogma of molecular biology.

Alternative splicing, or alternative RNA splicing, or differential splicing, is an alternative splicing process during gene expression that allows a single gene to code for multiple proteins. In this process, particular exons of a gene may be included within or excluded from the final, processed messenger RNA (mRNA) produced from that gene. This means the exons are joined in different combinations, leading to different (alternative) mRNA strands. Consequently, the proteins translated from alternatively spliced mRNAs usually contain differences in their amino acid sequence and, often, in their biological functions.

A protein isoform, or "protein variant", is a member of a set of highly similar proteins that originate from a single gene or gene family and are the result of genetic differences. While many perform the same or similar biological roles, some isoforms have unique functions. A set of protein isoforms may be formed from alternative splicings, variable promoter usage, or other post-transcriptional modifications of a single gene; post-translational modifications are generally not considered. Through RNA splicing mechanisms, mRNA has the ability to select different protein-coding segments (exons) of a gene, or even different parts of exons from RNA to form different mRNA sequences. Each unique sequence produces a specific form of a protein.

SR proteins are a conserved family of proteins involved in RNA splicing. SR proteins are named because they contain a protein domain with long repeats of serine and arginine amino acid residues, whose standard abbreviations are "S" and "R" respectively. SR proteins are ~200-600 amino acids in length and composed of two domains, the RNA recognition motif (RRM) region and the RS domain. SR proteins are more commonly found in the nucleus than the cytoplasm, but several SR proteins are known to shuttle between the nucleus and the cytoplasm.

A primary transcript is the single-stranded ribonucleic acid (RNA) product synthesized by transcription of DNA, and processed to yield various mature RNA products such as mRNAs, tRNAs, and rRNAs. The primary transcripts designated to be mRNAs are modified in preparation for translation. For example, a precursor mRNA (pre-mRNA) is a type of primary transcript that becomes a messenger RNA (mRNA) after processing.

Silent mutations are mutations in DNA that do not have an observable effect on the organism's phenotype. They are a specific type of neutral mutation. The phrase silent mutation is often used interchangeably with the phrase synonymous mutation; however, synonymous mutations are not always silent, nor vice versa. Synonymous mutations can affect transcription, splicing, mRNA transport, and translation, any of which could alter phenotype, rendering the synonymous mutation non-silent. The substrate specificity of the tRNA to the rare codon can affect the timing of translation, and in turn the co-translational folding of the protein. This is reflected in the codon usage bias that is observed in many species. Mutations that cause the altered codon to produce an amino acid with similar functionality are often classified as silent; if the properties of the amino acid are conserved, this mutation does not usually significantly affect protein function.
P elements are transposable elements that were discovered in Drosophila as the causative agents of genetic traits called hybrid dysgenesis. The transposon is responsible for the P trait of the P element and it is found only in wild flies. They are also found in many other eukaryotes.
Eukaryotic chromosome fine structure refers to the structure of sequences for eukaryotic chromosomes. Some fine sequences are included in more than one class, so the classification listed is not intended to be completely separate.
Exon shuffling is a molecular mechanism for the formation of new genes. It is a process through which two or more exons from different genes can be brought together ectopically, or the same exon can be duplicated, to create a new exon-intron structure. There are different mechanisms through which exon shuffling occurs: transposon mediated exon shuffling, crossover during sexual recombination of parental genomes and illegitimate recombination.

A splice site mutation is a genetic mutation that inserts, deletes or changes a number of nucleotides in the specific site at which splicing takes place during the processing of precursor messenger RNA into mature messenger RNA. Splice site consensus sequences that drive exon recognition are located at the very termini of introns. The deletion of the splicing site results in one or more introns remaining in mature mRNA and may lead to the production of abnormal proteins. When a splice site mutation occurs, the mRNA transcript possesses information from these introns that normally should not be included. Introns are supposed to be removed, while the exons are expressed.
In molecular biology, an exonic splicing enhancer (ESE) is a DNA sequence motif consisting of 6 bases within an exon that directs, or enhances, accurate splicing of heterogeneous nuclear RNA (hnRNA) or pre-mRNA into messenger RNA (mRNA).

Survival of motor neuron 1 (SMN1), also known as component of gems 1 or GEMIN1, is a gene that encodes the SMN protein in humans.

Survival of motor neuron 2 (SMN2) is a gene that encodes the SMN protein in humans.

HIKESHI is a protein important in lung and multicellular organismal development that, in humans, is encoded by the HIKESHI gene. HIKESHI is found on chromosome 11 in humans and chromosome 7 in mice. Similar sequences (orthologs) are found in most animal and fungal species. The mouse homolog, lethal gene on chromosome 7 Rinchik 6 protein is encoded by the l7Rn6 gene.
Splicing regulatory element (SRE) are cis-acting sequences in pre-mRNA, which either enhance or silence (suppress) the splicing of introns, or in general regulates the constitutive or alternative splicing of this pre-mRNA. SREs recruit trans-acting splicing factors to activate or suppress the splice site recognition or spliceosome assembly. The "context dependence" of SREs is categorized into at least two studied groups: (a) the location-dependent activity of SREs: the activity varies with the relative positions of SREs in pre-mRNA; (b) the gene-dependent activity of SREs: the SRE activity observed in one gene is lost when the SRE is moved to another gene.

Circular RNA is a type of single-stranded RNA which, unlike linear RNA, forms a covalently closed continuous loop. In circular RNA, the 3' and 5' ends normally present in an RNA molecule have been joined together. This feature confers numerous properties to circular RNA, many of which have only recently been identified.
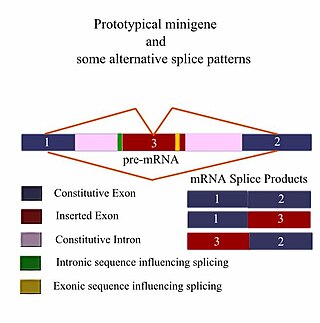
A minigene is a minimal gene fragment that includes an exon and the control regions necessary for the gene to express itself in the same way as a wild type gene fragment. This is a minigene in its most basic sense. More complex minigenes can be constructed containing multiple exons and intron(s). Minigenes provide a valuable tool for researchers evaluating splicing patterns both in vivo and in vitro biochemically assessed experiments. Specifically, minigenes are used as splice reporter vectors and act as a probe to determine which factors are important in splicing outcomes. They can be constructed to test the way both cis-regulatory elements and trans-regulatory elements affect gene expression.
Exitrons are produced through alternative splicing and have characteristics of both introns and exons, but are described as retained introns. Even though they are considered introns, which are typically cut out of pre mRNA sequences, there are significant problems that arise when exitrons are spliced out of these strands, with the most obvious result being altered protein structures and functions. They were first discovered in plants, but have recently been found in metazoan species as well.
References
- 1 2 Goren, Amir; Ram, Oren; Amit, Maayan; Keren, Hadas; Lev-Maor, Galit; Vig, Ida; Pupko, Tal; Ast, Gil (June 23, 2006). "Comparative Analysis Identifies Exonic Splicing Regulatory Sequences — The Complex Definition of Enhancers and Silencers". Molecular Cell. 22 (6): 769–81. doi: 10.1016/j.molcel.2006.05.008 . PMID 16793546.
- 1 2 Wang, Zefeng; Xiao, Xinshu; Van Nostrand, Eric; Burge, Christopher B. (2006-07-07). "General and Specific Functions of Exonic Splicing Silencers in Splicing Control". Molecular Cell. 23 (1): 61–70. doi:10.1016/j.molcel.2006.05.018. ISSN 1097-2765. PMC 1839040 . PMID 16797197.
- ↑ Souza, Jorge E. S. de; Ramalho, Rodrigo F.; Galante, Pedro A. F.; Meyer, Diogo; Souza, Sandro J. de (2011-07-01). "Alternative splicing and genetic diversity: silencers are more frequently modified by SNVs associated with alternative exon/intron borders". Nucleic Acids Research. 39 (12): 4942–4948. doi:10.1093/nar/gkr081. ISSN 0305-1048. PMC 3130264 . PMID 21398627.
- 1 2 3 4 5 6 7 8 9 Pozzoli, U.; Sironi, M. (2005-05-18). "Silencers regulate both constitutive and alternative splicing events in mammals". Cellular and Molecular Life Sciences. 62 (14): 1579–1604. doi:10.1007/s00018-005-5030-6. ISSN 1420-682X. PMID 15905961.