Related Research Articles

Malaria is a mosquito-borne infectious disease that affects humans and other vertebrates. Human malaria causes symptoms that typically include fever, fatigue, vomiting, and headaches. In severe cases, it can cause jaundice, seizures, coma, or death. Symptoms usually begin 10 to 15 days after being bitten by an infected mosquito. If not properly treated, people may have recurrences of the disease months later. In those who have recently survived an infection, reinfection usually causes milder symptoms. This partial resistance disappears over months to years if the person has no continuing exposure to malaria.

Plasmodium is a genus of unicellular eukaryotes that are obligate parasites of vertebrates and insects. The life cycles of Plasmodium species involve development in a blood-feeding insect host which then injects parasites into a vertebrate host during a blood meal. Parasites grow within a vertebrate body tissue before entering the bloodstream to infect red blood cells. The ensuing destruction of host red blood cells can result in malaria. During this infection, some parasites are picked up by a blood-feeding insect, continuing the life cycle.
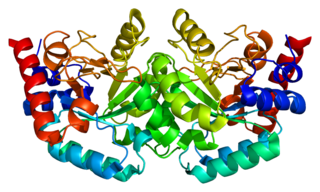
The enzyme Uridine monophosphate synthase catalyses the formation of uridine monophosphate (UMP), an energy-carrying molecule in many important biosynthetic pathways. In humans, the gene that codes for this enzyme is located on the long arm of chromosome 3 (3q13).

Plasmodium falciparum is a unicellular protozoan parasite of humans, and the deadliest species of Plasmodium that causes malaria in humans. The parasite is transmitted through the bite of a female Anopheles mosquito and causes the disease's most dangerous form, falciparum malaria. It is responsible for around 50% of all malaria cases. P. falciparum is therefore regarded as the deadliest parasite in humans. It is also associated with the development of blood cancer and is classified as a Group 2A (probable) carcinogen.

CD36, also known as platelet glycoprotein 4, fatty acid translocase (FAT), scavenger receptor class B member 3 (SCARB3), and glycoproteins 88 (GP88), IIIb (GPIIIB), or IV (GPIV) is a protein that in humans is encoded by the CD36 gene. The CD36 antigen is an integral membrane protein found on the surface of many cell types in vertebrate animals. It imports fatty acids inside cells and is a member of the class B scavenger receptor family of cell surface proteins. CD36 binds many ligands including collagen, thrombospondin, erythrocytes parasitized with Plasmodium falciparum, oxidized low density lipoprotein, native lipoproteins, oxidized phospholipids, and long-chain fatty acids.
Antigenic variation or antigenic alteration refers to the mechanism by which an infectious agent such as a protozoan, bacterium or virus alters the proteins or carbohydrates on its surface and thus avoids a host immune response, making it one of the mechanisms of antigenic escape. It is related to phase variation. Antigenic variation not only enables the pathogen to avoid the immune response in its current host, but also allows re-infection of previously infected hosts. Immunity to re-infection is based on recognition of the antigens carried by the pathogen, which are "remembered" by the acquired immune response. If the pathogen's dominant antigen can be altered, the pathogen can then evade the host's acquired immune system. Antigenic variation can occur by altering a variety of surface molecules including proteins and carbohydrates. Antigenic variation can result from gene conversion, site-specific DNA inversions, hypermutation, or recombination of sequence cassettes. The result is that even a clonal population of pathogens expresses a heterogeneous phenotype. Many of the proteins known to show antigenic or phase variation are related to virulence.
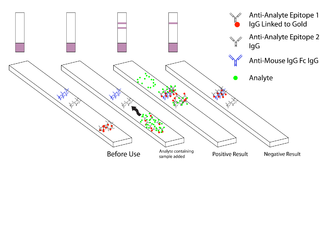
Malaria antigen detection tests are a group of commercially available rapid diagnostic tests of the rapid antigen test type that allow quick diagnosis of malaria by people who are not otherwise skilled in traditional laboratory techniques for diagnosing malaria or in situations where such equipment is not available. There are currently over 20 such tests commercially available. The first malaria antigen suitable as target for such a test was a soluble glycolytic enzyme Glutamate dehydrogenase. None of the rapid tests are currently as sensitive as a thick blood film, nor as cheap. A major drawback in the use of all current dipstick methods is that the result is essentially qualitative. In many endemic areas of tropical Africa, however, the quantitative assessment of parasitaemia is important, as a large percentage of the population will test positive in any qualitative assay.

Fusion proteins or chimeric (kī-ˈmir-ik) proteins are proteins created through the joining of two or more genes that originally coded for separate proteins. Translation of this fusion gene results in a single or multiple polypeptides with functional properties derived from each of the original proteins. Recombinant fusion proteins are created artificially by recombinant DNA technology for use in biological research or therapeutics. Chimeric or chimera usually designate hybrid proteins made of polypeptides having different functions or physico-chemical patterns. Chimeric mutant proteins occur naturally when a complex mutation, such as a chromosomal translocation, tandem duplication, or retrotransposition creates a novel coding sequence containing parts of the coding sequences from two different genes. Naturally occurring fusion proteins are commonly found in cancer cells, where they may function as oncoproteins. The bcr-abl fusion protein is a well-known example of an oncogenic fusion protein, and is considered to be the primary oncogenic driver of chronic myelogenous leukemia.
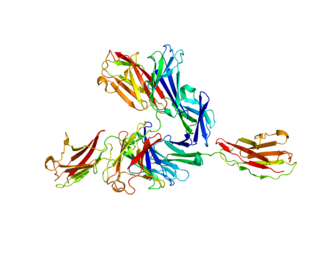
Basigin (BSG) also known as extracellular matrix metalloproteinase inducer (EMMPRIN) or cluster of differentiation 147 (CD147) is a protein that in humans is encoded by the BSG gene. This protein is a determinant for the Ok blood group system. There are three known antigens in the Ok system; the most common being Oka, OK2 and OK3. Basigin has been shown to be an essential receptor on red blood cells for the human malaria parasite, Plasmodium falciparum. The common isoform of basigin (basigin-2) has two immunoglobulin domains, and the extended form basigin-1 has three.
Malaria vaccines are vaccines that prevent malaria, a mosquito-borne infectious disease which annually affects an estimated 247 million people worldwide and causes 619,000 deaths. The first approved vaccine for malaria is RTS,S, known by the brand name Mosquirix. As of April 2023, the vaccine has been given to 1.5 million children living in areas with moderate-to-high malaria transmission. It requires at least three doses in infants by age 2, and a fourth dose extends the protection for another 1–2 years. The vaccine reduces hospital admissions from severe malaria by around 30%.
Plasmodium molecular tools are a set of methods for the genetic manipulation of the parasite genus Plasmodium. Plasmodium species have been difficult to scientifically study, partially due to the inability of many standard biological techniques to genetically alter the organism. Recent research has sought to overcome these technical barriers in order to make the parasite more amenable to study. Below is a description of published methods of genetic control within the Plasmodium parasite.
Human genetic resistance to malaria refers to inherited changes in the DNA of humans which increase resistance to malaria and result in increased survival of individuals with those genetic changes. The existence of these genotypes is likely due to evolutionary pressure exerted by parasites of the genus Plasmodium which cause malaria. Since malaria infects red blood cells, these genetic changes are most common alterations to molecules essential for red blood cell function, such as hemoglobin or other cellular proteins or enzymes of red blood cells. These alterations generally protect red blood cells from invasion by Plasmodium parasites or replication of parasites within the red blood cell.
Pregnancy-associated malaria (PAM) or placental malaria is a presentation of the common illness that is particularly life-threatening to both mother and developing fetus. PAM is caused primarily by infection with Plasmodium falciparum, the most dangerous of the four species of malaria-causing parasites that infect humans. During pregnancy, a woman faces a much higher risk of contracting malaria and of associated complications. Prevention and treatment of malaria are essential components of prenatal care in areas where the parasite is endemic – tropical and subtropical geographic areas. Placental malaria has also been demonstrated to occur in animal models, including in rodent and non-human primate models.

RTS,S/AS01 is a recombinant protein-based malaria vaccine. It is the only malaria vaccine approved and in wide use. As of April 2022, the vaccine has been given to 1 million children living in areas with moderate-to-high malaria transmission, with millions more doses to be provided as the vaccine's production expands. It requires at least three doses in infants by age 2, with a fourth dose extending the protection for another 1–2 years. The vaccine reduces hospital admissions from severe malaria by around 30%.
The AP2-Coincident Domain mainly at the Carboxy-terminus, or ACDC domain, is a protein domain that occurs in proteins from apicomplexan parasites.
The partial cleavage stimulation factor domain, or partial CstF domain, is a protein domain that occurs in proteins from apicomplexan parasites.
Circumsporozoite protein (CSP) is a secreted protein of the sporozoite stage of the malaria parasite and is the antigenic target of RTS,S and other malaria vaccines. The amino-acid sequence of CSP consists of an immunodominant central repeat region flanked by conserved motifs at the N- and C- termini that are implicated in protein processing as the parasite travels from the mosquito to the mammalian vector. The amino acid sequence of CSP was determined in 1984.
Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) is a family of proteins present on the membrane surface of red blood cells that are infected by the malarial parasite Plasmodium falciparum. PfEMP1 is synthesized during the parasite's blood stage inside the RBC, during which the clinical symptoms of falciparum malaria are manifested. Acting as both an antigen and adhesion protein, it is thought to play a key role in the high level of virulence associated with P. falciparum. It was discovered in 1984 when it was reported that infected RBCs had unusually large-sized cell membrane proteins, and these proteins had antibody-binding (antigenic) properties. An elusive protein, its chemical structure and molecular properties were revealed only after a decade, in 1995. It is now established that there is not one but a large family of PfEMP1 proteins, genetically regulated (encoded) by a group of about 60 genes called var. Each P. falciparum is able to switch on and off specific var genes to produce a functionally different protein, thereby evading the host's immune system. RBCs carrying PfEMP1 on their surface stick to endothelial cells, which facilitates further binding with uninfected RBCs, ultimately helping the parasite to both spread to other RBCs as well as bringing about the fatal symptoms of P. falciparum malaria.

The Plasmodium helical interspersed subtelomeric proteins (PHIST) or ring-infected erythrocyte surface antigens (RESA) are a family of protein domains found in the malaria-causing Plasmodium species. It was initially identified as a short four-helical conserved region in the single-domain export proteins, but the identification of this part associated with a DnaJ domain in P. falciparum RESA has led to its reclassification as the RESA N-terminal domain. This domain has been classified into three subfamilies, PHISTa, PHISTb, and PHISTc.
Reticulocyte binding protein homologs (RHs) are a superfamily of proteins found in Plasmodium responsible for cell invasion. Together with the family of erythrocyte binding-like proteins (EBLs) they make up the two families of invasion proteins universal to Plasmodium. The two families function cooperatively.
References
- 1 2 Oehring SC, Woodcroft BJ, Moes S, Wetzel J, Dietz O, Pulfer A, Dekiwadia C, Maeser P, Flueck C, Witmer K, Brancucci NM, Niederwieser I, Jenoe P, Ralph SA, Voss TS (November 2012). "Organellar proteomics reveals hundreds of novel nuclear proteins in the malaria parasite Plasmodium falciparum". Genome Biology. 13 (11): R108. doi:10.1186/gb-2012-13-11-r108. PMC 4053738 . PMID 23181666.