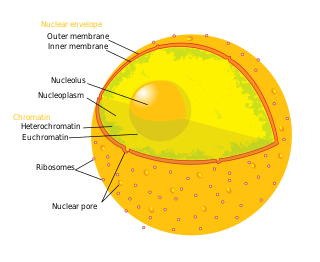
The nucleolus is the largest structure in the nucleus of eukaryotic cells. It is best known as the site of ribosome biogenesis, which is the synthesis of ribosomes. The nucleolus also participates in the formation of signal recognition particles and plays a role in the cell's response to stress. Nucleoli are made of proteins, DNA and RNA, and form around specific chromosomal regions called nucleolar organizing regions. Malfunction of nucleoli can be the cause of several human conditions called "nucleolopathies" and the nucleolus is being investigated as a target for cancer chemotherapy.

A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria; however, plasmids are sometimes present in archaea and eukaryotic organisms. In nature, plasmids often carry genes that benefit the survival of the organism and confer selective advantage such as antibiotic resistance. While chromosomes are large and contain all the essential genetic information for living under normal conditions, plasmids are usually very small and contain only additional genes that may be useful in certain situations or conditions. Artificial plasmids are widely used as vectors in molecular cloning, serving to drive the replication of recombinant DNA sequences within host organisms. In the laboratory, plasmids may be introduced into a cell via transformation. Synthetic plasmids are available for procurement over the internet.

Senescence or biological aging is the gradual deterioration of functional characteristics in living organisms. The word senescence can refer to either cellular senescence or to senescence of the whole organism. Organismal senescence involves an increase in death rates and/or a decrease in fecundity with increasing age, at least in the later part of an organism's life cycle. but it can be delayed. The 1934 discovery that calorie restriction can extend lifespans by 50% in rats, the existence of species having negligible senescence, and the existence of potentially immortal organisms such as members of the genus Hydra have motivated research into delaying senescence and thus age-related diseases. Rare human mutations can cause accelerated aging diseases.

Yeast artificial chromosomes (YACs) are genetically engineered chromosomes derived from the DNA of the yeast, Saccharomyces cerevisiae, which is then ligated into a bacterial plasmid. By inserting large fragments of DNA, from 100–1000 kb, the inserted sequences can be cloned and physically mapped using a process called chromosome walking. This is the process that was initially used for the Human Genome Project, however due to stability issues, YACs were abandoned for the use of bacterial artificial chromosome
RecQ helicase is a family of helicase enzymes initially found in Escherichia coli that has been shown to be important in genome maintenance. They function through catalyzing the reaction ATP + H2O → ADP + P and thus driving the unwinding of paired DNA and translocating in the 3' to 5' direction. These enzymes can also drive the reaction NTP + H2O → NDP + P to drive the unwinding of either DNA or RNA.
Double minutes are small fragments of extrachromosomal DNA, which have been observed in a large number of human tumors including breast, lung, ovary, colon, and most notably, neuroblastoma. They are a manifestation of gene amplification as a result of chromothripsis, during the development of tumors, which give the cells selective advantages for growth and survival. This selective advantage is as a result of double minutes frequently harboring amplified oncogenes and genes involved in drug resistance. DMs, like actual chromosomes, are composed of chromatin and replicate in the nucleus of the cell during cell division. Unlike typical chromosomes, they are composed of circular fragments of DNA, up to only a few million base pairs in size, and contain no centromere or telomere. Further to this, they often lack key regulatory elements, allowing genes to be constitutively expressed. The term ecDNA may be used to refer to DMs in a more general manner.
Extrachromosomal DNA is any DNA that is found off the chromosomes, either inside or outside the nucleus of a cell. Most DNA in an individual genome is found in chromosomes contained in the nucleus. Multiple forms of extrachromosomal DNA exist, and, while some of these serve important biological functions, they can also play a role in diseases such as cancer.
Sirtuins are a family of signaling proteins involved in metabolic regulation. They are ancient in animal evolution and appear to possess a highly conserved structure throughout all kingdoms of life. Chemically, sirtuins are a class of proteins that possess either mono-ADP-ribosyltransferase or deacylase activity, including deacetylase, desuccinylase, demalonylase, demyristoylase and depalmitoylase activity. The name Sir2 comes from the yeast gene 'silent mating-type information regulation 2', the gene responsible for cellular regulation in yeast.

DNA ligase 4 is an enzyme that in humans is encoded by the LIG4 gene.

Bloom syndrome protein is a protein that in humans is encoded by the BLM gene and is not expressed in Bloom syndrome.

Sirtuin 1, also known as NAD-dependent deacetylase sirtuin-1, is a protein that in humans is encoded by the SIRT1 gene.
Activated RNA polymerase II transcriptional coactivator p15 also known as positive cofactor 4 (PC4) or SUB1 homolog is a protein that in humans is encoded by the SUB1 gene. The human SUB1 gene is named after an orthologous gene in yeast.

NHP2-like protein 1 is a protein that in humans is encoded by the SNU13 gene.

Biomarkers of aging are biomarkers that could predict functional capacity at some later age better than chronological age. Stated another way, biomarkers of aging would give the true "biological age", which may be different from the chronological age.
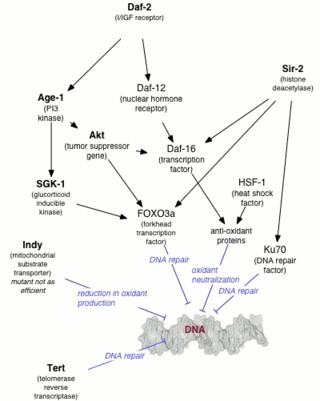
Genetics of aging is generally concerned with life extension associated with genetic alterations, rather than with accelerated aging diseases leading to reduction in lifespan.

An R-loop is a three-stranded nucleic acid structure, composed of a DNA:RNA hybrid and the associated non-template single-stranded DNA. R-loops may be formed in a variety of circumstances and may be tolerated or cleared by cellular components. The term "R-loop" was given to reflect the similarity of these structures to D-loops; the "R" in this case represents the involvement of an RNA moiety.
Extrachromosomal circular DNA (eccDNA) is a type of double-stranded circular DNA structure that was first discovered in 1964 by Alix Bassel and Yasuo Hotta. In contrast to previously identified circular DNA structures, eccDNA are circular DNA found in the eukaryotic nuclei of plant and animal cells. Extrachromosomal circular DNA is derived from chromosomal DNA, can range in size from 50 base pairs to several mega-base pairs in length, and can encode regulatory elements and full-length genes. eccDNA has been observed in various eukaryotic species and it is proposed to be a byproduct of programmed DNA recombination events, such as V(D)J recombination.

RRM3 is a gene that encodes a 5′-to-3′ DNA helicase known affect multiple cellular replication and repair processes and is most commonly studied in Saccharomyces cerevisiae. RRM3 formally stands for Ribosomal DNArecombination mutation 3. The gene codes for nuclear protein Rrm3p, which is 723 amino acids in length, and is part of a Pif1p DNA helicase sub-family that is conserved from yeasts to humans. RRM3 and its encoded protein have been shown to be vital for cellular replication, specifically associating with replication forks genome-wide. RRM3 is located on chromosome 8 in yeast cells and codes for 723 amino acids producing a protein that weighs 81,581 Da.

MicroDNA is the most abundant subtype of Extrachromosomal Circular DNA (eccDNA) in humans, typically ranging from 200-400 base pairs in length and enriched in non-repetitive genomic sequences with a high density of exons. Additionally, microDNA has been found to come from regions with CpG-islands which are commonly found within the 5' and 3' UTRs. Being produced from regions of active transcription, it is hypothesized that microDNA may be formed as a by-product of transcriptional DNA damage repair. MicroDNA is also thought to arise from other DNA repair pathways, mainly due to the parental sequences of microDNA having 2- to 15 bp direct repeats at the ends, resulting in replication slippage repair. While only recently discovered, the role microDNA plays in and out of the cell is still not completely understood. However, microDNA is currently thought to affect cellular homeostasis through transcription factor binding and have been used as a cancer biomarker.
Anindya Dutta is an Indian-born American biochemist and cancer researcher, a Chair of the Department of Genetics at the University of Alabama at Birmingham School of Medicine since 2021, who has served as Chair of the Department of Biochemistry and Molecular Genetics at the University of Virginia School of Medicine in 2011–2021. Dutta's research has focused on the mammalian cell cycle with an emphasis on DNA replication and repair and on noncoding RNAs. He is particularly interested in how de-regulation of these processes promote cancer progression. For his accomplishments he has been elected a Fellow of the American Association for the Advancement of Science, received the Ranbaxy Award in Biomedical Sciences, the Outstanding Investigator Award from the American Society for Investigative Pathology, the Distinguished Scientist Award from the University of Virginia and the Mark Brothers Award from the Indiana University School of Medicine.














