Related Research Articles

Nanoarchaeota is a proposed phylum in the domain Archaea that currently has only one representative, Nanoarchaeum equitans, which was discovered in a submarine hydrothermal vent and first described in 2002.

The Korarchaeota is a proposed phylum within the Archaea. The name is derived from the Greek noun koros or kore, meaning young man or young woman, and the Greek adjective archaios which means ancient. They are also known as Xenarchaeota. The name is equivalent to Candidatus Korarchaeota, and they go by the name Xenarchaeota or Xenarchaea as well.
The Aquificota phylum is a diverse collection of bacteria that live in harsh environmental settings. The name Aquificota was given to this phylum based on an early genus identified within this group, Aquifex, which is able to produce water by oxidizing hydrogen. They have been found in springs, pools, and oceans. They are autotrophs, and are the primary carbon fixers in their environments. These bacteria are Gram-negative, non-spore-forming rods. They are true bacteria as opposed to the other inhabitants of extreme environments, the Archaea.
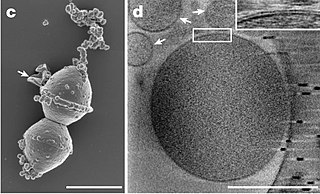
In prokaryote nomenclature, Candidatus is used to name prokaryotic taxa that are well characterized but yet-uncultured. Contemporary sequencing approaches, such as 16S ribosomal RNA sequencing or metagenomics, provide much information about the analyzed organisms and thus allow identification and characterization of individual species. However, the majority of prokaryotic species remain uncultivable and hence inaccessible for further characterization in in vitro study. The recent discoveries of a multitude of candidate taxa has led to candidate phyla radiation expanding the tree of life through the new insights in bacterial diversity.

Halomonadaceae is a family of halophilic Pseudomonadota.

Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribosomal DNA (rDNA) and then bound to ribosomal proteins to form small and large ribosome subunits. rRNA is the physical and mechanical factor of the ribosome that forces transfer RNA (tRNA) and messenger RNA (mRNA) to process and translate the latter into proteins. Ribosomal RNA is the predominant form of RNA found in most cells; it makes up about 80% of cellular RNA despite never being translated into proteins itself. Ribosomes are composed of approximately 60% rRNA and 40% ribosomal proteins, though this ratio differs between prokaryotes and eukaryotes.
The Clostridiaceae are a family of the bacterial class Clostridia, and contain the genus Clostridium.
A structural gene is a gene that codes for any RNA or protein product other than a regulatory factor. A term derived from the lac operon, structural genes are typically viewed as those containing sequences of DNA corresponding to the amino acids of a protein that will be produced, as long as said protein does not function to regulate gene expression. Structural gene products include enzymes and structural proteins. Also encoded by structural genes are non-coding RNAs, such as rRNAs and tRNAs.
Parachlamydiaceae is a family of bacteria in the order Chlamydiales. Species in this family have a Chlamydia–like cycle of replication and their ribosomal RNA genes are 80–90% identical to ribosomal genes in the Chlamydiaceae. The Parachlamydiaceae naturally infect amoebae and can be grown in cultured Vero cells. The Parachlamydiaceae are not recognized by monoclonal antibodies that detect Chlamydiaceae lipopolysaccharide.

16S ribosomal RNA is the RNA component of the 30S subunit of a prokaryotic ribosome. It binds to the Shine-Dalgarno sequence and provides most of the SSU structure.
Lentisphaerota is a phylum of bacteria closely related to Chlamydiota and Verrucomicrobiota.
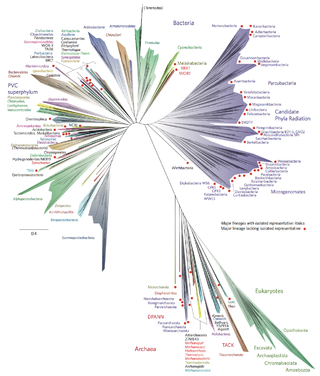
Bacterial phyla constitute the major lineages of the domain Bacteria. While the exact definition of a bacterial phylum is debated, a popular definition is that a bacterial phylum is a monophyletic lineage of bacteria whose 16S rRNA genes share a pairwise sequence identity of ~75% or less with those of the members of other bacterial phyla.

Bacterial taxonomy is subfield of taxonomy devoted to the classification of bacteria specimens into taxonomic ranks.
Armatimonadota is a phylum of gram-negative bacteria.
Beutenbergiaceae is an Actinomycete family.
Thermomonosporaceae is a family of bacteria that share similar genotypic and phenotypic characteristics. The family Thermomonosporaceae includes aerobic, Gram-positive, non-acid-fast, chemo-organotrophic Actinomycetota. They produce a branched substrate mycelium bearing aerial hyphae that undergo differentiation into single or short chains of arthrospores. All species of Thermomonosporaceae share the same cell wall type, a similar menaquinone profile in which MK-9(H6)is predominant, and fatty acid profile type 3a. The presence of the diagnostic sugar madurose is variable, but can be found in most species of this family. The polar lipid profiles are characterized as phospholipid type PI for most species of Thermomonospora, Actinomadura and Spirillospora. The members of Actinocorallia are characterized by phospholipid type PII.
Microbial DNA barcoding is the use of DNA metabarcoding to characterize a mixture of microorganisms. DNA metabarcoding is a method of DNA barcoding that uses universal genetic markers to identify DNA of a mixture of organisms.

Thermodesulfobacterium hveragerdense is a bacterial species belonging to genus Thermodesulfobacterium, which are thermophilic sulfate-reducing bacteria. This species is found in aquatic areas of high temperature, and lives in freshwater like most, but not all Thermodesulfobacterium species It was first isolated from hotsprings in Iceland.
Natrialbales is an order of halophilic, chemoorganotrophic archaea within the class Haloarchaea. The type genus of this order is Natrialba.
Haloferacales is an order of halophilic, chemoorganotrophic or heterotrophic archaea within the class Haloarchaea. The type genus of this order is Haloferax.
References
- ↑ Chun, Jongsik; Lee, Jae-Hak; Jung, Yoonyoung; Kim, Myungjin; Kim, Seil; Kwon Kim, Byung; Lim, Young-Woon (2007). "EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences". International Journal of Systematic and Evolutionary Microbiology. 57 (10): 2259–2261. doi: 10.1099/ijs.0.64915-0 . PMID 17911292.
- ↑ Kim, OS; Cho, YJ; Lee, K; Yoon, SH; Kim, M; Na, H; Park, SC; Jeon, YS; Lee, JH; Yi, H; Won, S; Chun, J (2012). "Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species". Int J Syst Evol Microbiol. 62 (3): 716–21. doi:10.1099/ijs.0.038075-0. PMID 22140171.