A dehydrogenase (also called DH or DHase in the literature) is an enzyme belonging to the group of oxidoreductases that oxidizes a substrate by reducing an electron acceptor, usually NAD+/NADP+ or a flavin coenzyme such as FAD or FMN. They also catalyze the reverse reaction, for instance alcohol dehydrogenase not only oxidizes ethanol to acetaldehyde in animals but also produces ethanol from acetaldehyde in yeast.

Flavin is the common name for a group of organic compounds based on pteridine, formed by the tricyclic heterocycle isoalloxazine. The biochemical source is the vitamin riboflavin. The flavin moiety is often attached with an adenosine diphosphate to form flavin adenine dinucleotide (FAD), and, in other circumstances, is found as flavin mononucleotide, a phosphorylated form of riboflavin. It is in one or the other of these forms that flavin is present as a prosthetic group in flavoproteins.

The glucose oxidase enzyme also known as notatin is an oxido-reductase that catalyses the oxidation of glucose to hydrogen peroxide and D-glucono-δ-lactone. This enzyme is produced by certain species of fungi and insects and displays antibacterial activity when oxygen and glucose are present.

In biochemistry, flavin adenine dinucleotide (FAD) is a redox-active coenzyme associated with various proteins, which is involved with several enzymatic reactions in metabolism. A flavoprotein is a protein that contains a flavin group, which may be in the form of FAD or flavin mononucleotide (FMN). Many flavoproteins are known: components of the succinate dehydrogenase complex, α-ketoglutarate dehydrogenase, and a component of the pyruvate dehydrogenase complex.
Catechol oxidase is a copper oxidase that contains a type 3 di-copper cofactor and catalyzes the oxidation of ortho-diphenols into ortho-quinones coupled with the reduction of molecular oxygen to water. It is present in a variety of species of plants and fungi including Ipomoea batatas and Camellia sinensis. Metalloenzymes with type 3 copper centers are characterized by their ability to reversibly bind dioxygen at ambient conditions. In plants, catechol oxidase plays a key role in enzymatic browning by catalyzing the oxidation of catechol to o-quinone in the presence of oxygen, which can rapidly polymerize to form the melanin that grants damaged fruits their dark brown coloration.
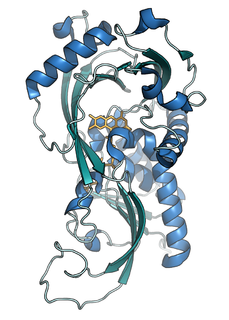
D-amino acid oxidase is an enzyme with the function on a molecular level to oxidize D-amino acids to the corresponding α-keto acids, producing ammonia and hydrogen peroxide. This results in a number of physiological effects in various systems, most notably the brain. The enzyme is most active toward neutral D-amino acids, and not active toward acidic D-amino acids. One of its most important targets in mammals is D-Serine in the central nervous system. By targeting this and other D-amino acids in vertebrates, DAAO is important in detoxification. The role in microorganisms is slightly different, breaking down D-amino acids to generate energy.
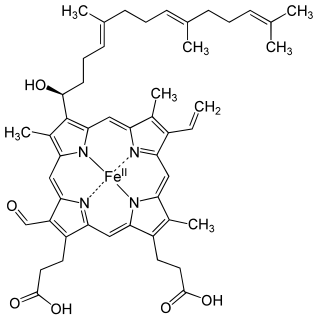
Heme A is a heme, a coordination complex consisting of a macrocyclic ligand called a porphyrin, chelating an iron atom. Heme A is a biomolecule and is produced naturally by many organisms. Heme A, often appears a dichroic green/red when in solution, is a structural relative of heme B, a component of hemoglobin, the red pigment in blood.
In enzymology, sarcosine dehydrogenase (EC 1.5.8.3) is a mitochondrial enzyme that catalyzes the chemical reaction N-demethylation of sarcosine to give glycine. This enzyme belongs to the family of oxidoreductases, specifically those acting on the CH-NH group of donor with other acceptors. The systematic name of this enzyme class is sarcosine:acceptor oxidoreductase (demethylating). Other names in common use include sarcosine N-demethylase, monomethylglycine dehydrogenase, and sarcosine:(acceptor) oxidoreductase (demethylating). Sarcosine dehydrogenase is closely related to dimethylglycine dehydrogenase, which catalyzes the demethylation reaction of dimethylglycine to sarcosine. Both sarcosine dehydrogenase and dimethylglycine dehydrogenase use FAD as a cofactor. Sarcosine dehydrogenase is linked by electron-transferring flavoprotein (ETF) to the respiratory redox chain. The general chemical reaction catalyzed by sarcosine dehydrogenase is:
In enzymology, an acyl-CoA oxidase (EC 1.3.3.6) is an enzyme that catalyzes the chemical reaction
In enzymology, a L-galactonolactone oxidase (EC 1.3.3.12) is an enzyme that catalyzes the chemical reaction
In enzymology, a malate oxidase (EC 1.1.3.3) is an enzyme that catalyzes the chemical reaction
In enzymology, a vanillyl-alcohol oxidase (EC 1.1.3.38) is an enzyme that catalyzes the chemical reaction
In enzymology, a pyruvate oxidase (EC 1.2.3.3) is an enzyme that catalyzes the chemical reaction
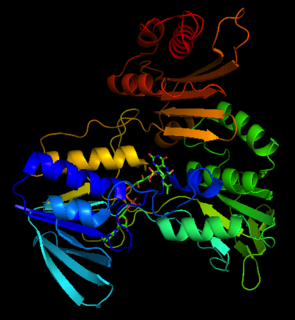
In enzymology, a NADH peroxidase (EC 1.11.1.1) is an enzyme that catalyzes the chemical reaction

Amine oxidase (copper-containing) (AOC) (EC 1.4.3.21 and EC 1.4.3.22; formerly EC 1.4.3.6) is a family of amine oxidase enzymes which includes both primary-amine oxidase and diamine oxidase; these enzymes catalyze the oxidation of a wide range of biogenic amines including many neurotransmitters, histamine and xenobiotic amines. They act as a disulphide-linked homodimer. They catalyse the oxidation of primary amines to aldehydes, with the subsequent release of ammonia and hydrogen peroxide, which requires one copper ion per subunit and topaquinone as cofactor:
In enzymology, a D-aspartate oxidase (EC 1.4.3.1) is an enzyme that catalyzes the chemical reaction
In enzymology, a putrescine oxidase (EC 1.4.3.10) is an enzyme that catalyzes the chemical reaction

In molecular biology, the FAD dependent oxidoreductase family of proteins is a family of FAD dependent oxidoreductases. Members of this family include Glycerol-3-phosphate dehydrogenase EC 1.1.99.5, Sarcosine oxidase beta subunit EC 1.5.3.1, D-amino-acid dehydrogenase EC 1.4.99.1, D-aspartate oxidase EC 1.4.3.1.

Tetrahydrocannabinolic acid (THCA) synthase is an enzyme responsible for catalyzing the formation of THCA from cannabigerolic acid (CBGA). THCA is the direct precursor of tetrahydrocannabinol (THC), the principal psychoactive component of cannabis, which is produced from various strains of Cannabis sativa. Therefore, THCA synthase is considered to be a key enzyme controlling cannabis psychoactivity. Polymorphisms of THCA synthase result in varying levels of THC in Cannabis plants, resulting in "drug-type" and "fiber-type" C. sativa varieties.

Galactose oxidase is an enzyme that catalyzes the oxidation of D-galactose in some species of fungi.










