
Nanoarchaeota is a proposed phylum in the domain Archaea that currently has only one representative, Nanoarchaeum equitans, which was discovered in a submarine hydrothermal vent and first described in 2002.
The Aquificota phylum is a diverse collection of bacteria that live in harsh environmental settings. The name Aquificota was given to this phylum based on an early genus identified within this group, Aquifex, which is able to produce water by oxidizing hydrogen. They have been found in springs, pools, and oceans. They are autotrophs, and are the primary carbon fixers in their environments. These bacteria are Gram-negative, non-spore-forming rods. They are true bacteria as opposed to the other inhabitants of extreme environments, the Archaea.
Pseudomonas citronellolis is a Gram-negative, bacillus bacterium that is used to study the mechanisms of pyruvate carboxylase. It was first isolated from forest soil, under pine trees, in northern Virginia, United States.

Archaeal Richmond Mine acidophilic nanoorganisms (ARMAN) were first discovered in an extremely acidic mine located in northern California (Richmond Mine at Iron Mountain) by Brett Baker in Jill Banfield's laboratory at the University of California Berkeley. These novel groups of archaea named ARMAN-1, ARMAN-2 (Candidatus Micrarchaeum acidiphilum ARMAN-2), and ARMAN-3 were missed by previous PCR-based surveys of the mine community because the ARMANs have several mismatches with commonly used PCR primers for 16S rRNA genes. Baker et al. detected them in a later study using shotgun sequencing of the community. The three groups were originally thought to represent three unique lineages deeply branched within the Euryarchaeota, a subgroup of the Archaea. However, based on a more complete archaeal genomic tree, they were assigned to a new superphylum named DPANN. The ARMAN groups now comprise deeply divergent phyla named Micrarchaeota and Parvarchaeota. Their 16S rRNA genes differ by as much as 17% between the three groups. Prior to their discovery, all of the Archaea shown to be associated with Iron Mountain belonged to the order Thermoplasmatales (e.g., Ferroplasma acidarmanus).

16S ribosomal RNA is the RNA component of the 30S subunit of a prokaryotic ribosome. It binds to the Shine-Dalgarno sequence and provides most of the SSU structure.
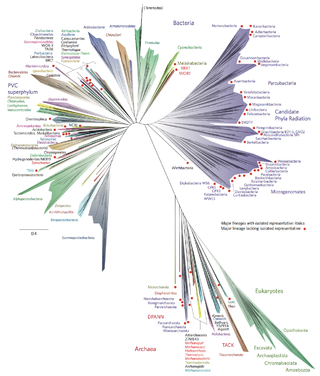
Bacterial phyla constitute the major lineages of the domain Bacteria. While the exact definition of a bacterial phylum is debated, a popular definition is that a bacterial phylum is a monophyletic lineage of bacteria whose 16S rRNA genes share a pairwise sequence identity of ~75% or less with those of the members of other bacterial phyla.
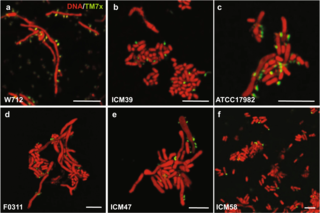
Saccharibacteria, formerly known as TM7, is a major bacterial lineage. It was discovered through 16S rRNA sequencing.
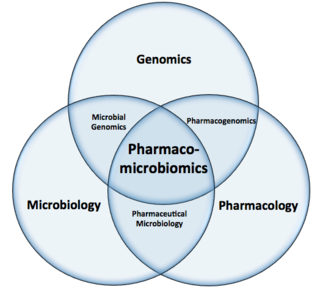
Pharmacomicrobiomics, proposed by Prof. Marco Candela for the ERC-2009-StG project call, and publicly coined for the first time in 2010 by Rizkallah et al., is defined as the effect of microbiome variations on drug disposition, action, and toxicity. Pharmacomicrobiomics is concerned with the interaction between xenobiotics, or foreign compounds, and the gut microbiome. It is estimated that over 100 trillion prokaryotes representing more than 1000 species reside in the gut. Within the gut, microbes help modulate developmental, immunological and nutrition host functions. The aggregate genome of microbes extends the metabolic capabilities of humans, allowing them to capture nutrients from diverse sources. Namely, through the secretion of enzymes that assist in the metabolism of chemicals foreign to the body, modification of liver and intestinal enzymes, and modulation of the expression of human metabolic genes, microbes can significantly impact the ingestion of xenobiotics.
The candidate phyla radiation is a large evolutionary radiation of bacterial lineages whose members are mostly uncultivated and only known from metagenomics and single cell sequencing. They have been described as nanobacteria or ultra-small bacteria due to their reduced size (nanometric) compared to other bacteria.
TM7x, also known as Nanosynbacter lyticus type strain TM7x HMT 952. is a phylotype of one of the most enigmatic phyla, Candidatus Saccharibacteria, formerly candidate phylum TM7. It is the only member of the candidate phylum that has been cultivated successfully from the human oral cavity, and stably maintained in vitro. and serves as a crucial paradigm. of the newly described Candidate Phyla Radiation (CPR). The cultivated oral taxon is designated as Saccharibacteria oral taxon TM7x. TM7x has a unique lifestyle in comparison to other bacteria that are associated with humans. It is an obligate epibiont parasite, or an "epiparasite", growing on the surface of its host bacterial species Actinomyces odontolyticus subspecies actinosynbacter strain XH001, which is referred to as the "basibiont". Actinomyces species are one of the early microbial colonizers in the oral cavity. Together, they exhibit parasitic epibiont symbiosis.
The Microgenomatota or Microgenomates are a proposed supergroup of bacterial candidate phyla in the Candidate Phyla Radiation.
Gracilibacteria is a bacterial candidate phylum formerly known as GN02, BD1-5, or SN-2. It is part of the Candidate Phyla Radiation and the Patescibacteria group.
Zixibacteria is a bacterial phylum with candidate status, meaning it had no cultured representatives. It is a member of the FCB group
Katanobacteria is a bacterial phylum formerly known as WWE3. It has candidate status, meaning there are no cultured representatives, and is a member of the Candidate Phyla Radiation (CPR).
Fermentibacteria is a bacterial phylum with candidate status. It is part of the FCB group.
Berkelbacteria is a bacterial phylum with candidate status, meaning there are no cultured representatives for this group. It is part of the Candidate Phyla Radiation.
Delphibacteria is a candidate bacterial phylum in the FCB group. The phylum was first proposed after analysis of two genomes from the mouths of two bottlenose dolphins. "Dephibacteria" was proposed in recognition of the first genomic representatives having been recovered from the dolphin mouth. Members of the Delphibacteria phylum have been retroactively detected in a variety of environments.
Kryptonia is a bacterial phylum with candidate status. It is a member of the FCB group.
Modulibacteria(Moduliflexota) is a bacterial phylum formerly known as KS3B3 or GN06. It is a candidate phylum, meaning there are no cultured representatives of this group. Members of the Modulibacteria phylum are known to cause fatal filament overgrowth (bulking) in high-rate industrial anaerobic wastewater treatment bioreactors.
There are several models of the branching order of bacterial phyla, one of these is the Genome Taxonomy Database (GTDB).






