Related Research Articles

Arabidopsis thaliana, the thale cress, mouse-ear cress or arabidopsis, is a small plant from the mustard family (Brassicaceae), native to Eurasia and Africa. Commonly found along the shoulders of roads and in disturbed land, it is generally considered a weed.

Vernalization is the induction of a plant's flowering process by exposure to the prolonged cold of winter, or by an artificial equivalent. After vernalization, plants have acquired the ability to flower, but they may require additional seasonal cues or weeks of growth before they will actually do so. The term is sometimes used to refer to the need of herbal (non-woody) plants for a period of cold dormancy in order to produce new shoots and leaves, but this usage is discouraged.
Gibberellins (GAs) are plant hormones that regulate various developmental processes, including stem elongation, germination, dormancy, flowering, flower development, and leaf and fruit senescence. GAs are one of the longest-known classes of plant hormone. It is thought that the selective breeding of crop strains that were deficient in GA synthesis was one of the key drivers of the "green revolution" in the 1960s, a revolution that is credited to have saved over a billion lives worldwide.

Antisense RNA (asRNA), also referred to as antisense transcript, natural antisense transcript (NAT) or antisense oligonucleotide, is a single stranded RNA that is complementary to a protein coding messenger RNA (mRNA) with which it hybridizes, and thereby blocks its translation into protein. The asRNAs have been found in both prokaryotes and eukaryotes, and can be classified into short and long non-coding RNAs (ncRNAs). The primary function of asRNA is regulating gene expression. asRNAs may also be produced synthetically and have found wide spread use as research tools for gene knockdown. They may also have therapeutic applications.
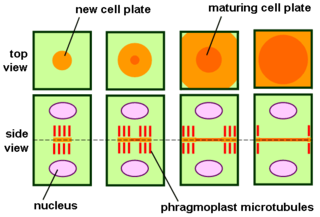
The phragmoplast is a plant cell specific structure that forms during late cytokinesis. It serves as a scaffold for cell plate assembly and subsequent formation of a new cell wall separating the two daughter cells. The phragmoplast can only be observed in Phragmoplastophyta, a clade that includes the Coleochaetophyceae, Zygnematophyceae, Mesotaeniaceae, and Embryophyta. Some algae use another type of microtubule array, a phycoplast, during cytokinesis.

In molecular genetics, a repressor is a DNA- or RNA-binding protein that inhibits the expression of one or more genes by binding to the operator or associated silencers. A DNA-binding repressor blocks the attachment of RNA polymerase to the promoter, thus preventing transcription of the genes into messenger RNA. An RNA-binding repressor binds to the mRNA and prevents translation of the mRNA into protein. This blocking or reducing of expression is called repression.
Florigen is the hypothesized hormone-like molecule responsible for controlling and/or triggering flowering in plants. Florigen is produced in the leaves, and acts in the shoot apical meristem of buds and growing tips. It is known to be graft-transmissible, and even functions between species. Florigen has been found identical to the transcription factor FLOWERING LOCUS T (FT).
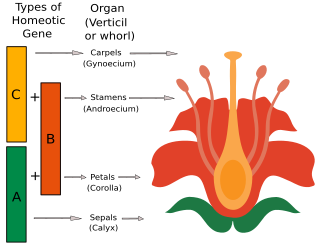
The ABC model of flower development is a scientific model of the process by which flowering plants produce a pattern of gene expression in meristems that leads to the appearance of an organ oriented towards sexual reproduction, a flower. There are three physiological developments that must occur in order for this to take place: firstly, the plant must pass from sexual immaturity into a sexually mature state ; secondly, the transformation of the apical meristem's function from a vegetative meristem into a floral meristem or inflorescence; and finally the growth of the flower's individual organs. The latter phase has been modelled using the ABC model, which aims to describe the biological basis of the process from the perspective of molecular and developmental genetics.
Polycomb-group proteins are a family of protein complexes first discovered in fruit flies that can remodel chromatin such that epigenetic silencing of genes takes place. Polycomb-group proteins are well known for silencing Hox genes through modulation of chromatin structure during embryonic development in fruit flies. They derive their name from the fact that the first sign of a decrease in PcG function is often a homeotic transformation of posterior legs towards anterior legs, which have a characteristic comb-like set of bristles.
The MADS box is a conserved sequence motif. The genes which contain this motif are called the MADS-box gene family. The MADS box encodes the DNA-binding MADS domain. The MADS domain binds to DNA sequences of high similarity to the motif CC[A/T]6GG termed the CArG-box. MADS-domain proteins are generally transcription factors. The length of the MADS-box reported by various researchers varies somewhat, but typical lengths are in the range of 168 to 180 base pairs, i.e. the encoded MADS domain has a length of 56 to 60 amino acids. There is evidence that the MADS domain evolved from a sequence stretch of a type II topoisomerase in a common ancestor of all extant eukaryotes.
Evolutionary developmental biology (evo-devo) is the study of developmental programs and patterns from an evolutionary perspective. It seeks to understand the various influences shaping the form and nature of life on the planet. Evo-devo arose as a separate branch of science rather recently. An early sign of this occurred in 1999.

Dame Caroline Dean is a British plant scientist working at the John Innes Centre. She is focused on understanding the molecular controls used by plants to seasonally judge when to flower. She is specifically interested in vernalisation — the acceleration of flowering in plants by exposure to periods of prolonged cold. She has also been on the Life Sciences jury for the Infosys Prize from 2018.
Martin Edward Kreitman is an American geneticist at the University of Chicago, most well known for the McDonald–Kreitman test that is used to infer the amount of adaptive evolution in population genetic studies.
Richard Arthur is a professor of biochemistry and genetics at the University of Wisconsin-Madison. He got his bachelor's degree in biology at Pennsylvania State University. He went on to receive his PhD in biochemistry at Indiana University in 1982 and did post doctoral research at the University of Washington. Amasino's research focuses on plants and how plants know when to flower. In 2006 he was elected to the National Academy of Sciences.
Carotene epsilon-monooxygenase (EC 1.14.99.45, CYP97C1, LUT1) is an enzyme with systematic name alpha-carotene:oxygen oxidoreductase (3-hydroxylating). This enzyme catalyses the following chemical reaction
Elizabeth Jean Finnegan FAA is an Australian botanist who researches plant flowering processes and epigenetic regulation in plants. She currently works at the Commonwealth Scientific and Industrial Research Organisation (CSIRO) as a senior scientist, leading research on the "Control of Floral Initiation", part of the CSIRO Agriculture Flagship.
Elizabeth Salisbury Dennis is an Australian scientist working mainly in the area of plant molecular biology. She is currently a chief scientist at the plant division of CSIRO Canberra. She was elected a Fellow of the Australian Academy of Technological Sciences and Engineering (FTSE) in 1987, and the Australian Academy of Science in 1995. She jointly received the inaugural Prime Minister's Science Prize together with Professor Jim Peacock in 2000 for her outstanding achievements in science and technology.

Camalexin (3-thiazol-2-yl-indole) is a simple indole alkaloid found in the plant Arabidopsis thaliana and other crucifers. The secondary metabolite functions as a phytoalexin to deter bacterial and fungal pathogens.
Plants depend on epigenetic processes for proper function. Epigenetics is defined as "the study of changes in gene function that are mitotically and/or meiotically heritable and that do not entail a change in DNA sequence". The area of study examines protein interactions with DNA and its associated components, including histones and various other modifications such as methylation, which alter the rate or target of transcription. Epi-alleles and epi-mutants, much like their genetic counterparts, describe changes in phenotypes due to epigenetic mechanisms. Epigenetics in plants has attracted scientific enthusiasm because of its importance in agriculture.

RNA-directed DNA methylation (RdDM) is a biological process in which non-coding RNA molecules direct the addition of DNA methylation to specific DNA sequences. The RdDM pathway is unique to plants, although other mechanisms of RNA-directed chromatin modification have also been described in fungi and animals. To date, the RdDM pathway is best characterized within angiosperms, and particularly within the model plant Arabidopsis thaliana. However, conserved RdDM pathway components and associated small RNAs (sRNAs) have also been found in other groups of plants, such as gymnosperms and ferns. The RdDM pathway closely resembles other sRNA pathways, particularly the highly conserved RNAi pathway found in fungi, plants, and animals. Both the RdDM and RNAi pathways produce sRNAs and involve conserved Argonaute, Dicer and RNA-dependent RNA polymerase proteins.
References
- ↑ Michaels SD, Amasino RM (1999), "FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering", Plant Cell, 11 (5): 949–56, doi:10.1105/tpc.11.5.949, PMC 144226 , PMID 10330478.
- ↑ Sheldon, Cc; Rouse, Dt; Finnegan, Ej; Peacock, Wj; Dennis, Es (Mar 2000), "The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC).", Proceedings of the National Academy of Sciences of the United States of America, 97 (7): 3753–8, doi: 10.1073/pnas.060023597 , PMC 16312 , PMID 10716723
- ↑ Lim, Mh; Kim, J; Kim, Ys; Chung, Ks; Seo, Yh; Lee, I; Kim, J; Hong, Cb; Kim, Hj; Park, Cm (Mar 2004), "A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS C.", The Plant Cell, 16 (3): 731–40, doi:10.1105/tpc.019331, PMC 385284 , PMID 14973162
- 1 2 Sheldon, C. C.; Conn, AB; Dennis, ES; Peacock, WJ (2002), "Different Regulatory Regions Are Required for the Vernalization-Induced Repression of FLOWERING LOCUS C and for the Epigenetic Maintenance of Repression", The Plant Cell Online, 14 (10): 2527–37, doi:10.1105/tpc.004564, PMC 151233 , PMID 12368502
- ↑ Michaels, Sd; He, Y; Scortecci, Kc; Amasino, Rm (Aug 2003), "Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis", Proceedings of the National Academy of Sciences of the United States of America, 100 (17): 10102–7, Bibcode:2003PNAS..10010102M, doi: 10.1073/pnas.1531467100 , PMC 187779 , PMID 12904584