
The Actino-pnp RNA motif is a conserved structure found in Actinomycetota that is apparently in the 5' untranslated regions of genes predicted to encode exoribonucleases. The RNA element's function is likely analogous to an RNA structure found upstream of polynucleotide phosphorylase genes in E. coli and related enterobacteria. In this latter system, the polynucleotide phosphorlyase gene regulates its own expression levels by a feedback mechanism that involves its activity upon the RNA structure. However, the E. coli RNA appears to be structurally unrelated to the Actino-pnp motif.
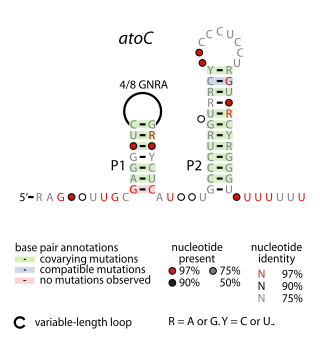
The atoC RNA motif is a conserved RNA-like structure identified by bioinformatics. It consistently appears upstream of protein-coding gene that are predicted to encode oxidoreductase activity, dihydropteroate synthase or DNA-binding response regulators.

The Bacteroid-trp RNA motif is a conserved RNA element detected by bioinformatics. It is found in the phylum Bacteroidota in the apparent 5' untranslated regions of genes that encode enzymes used in the synthesis of the amino acid tryptophan. A short open reading frame is found within the motif that encodes at least two tryptophan codons. Similar motifs have been identified regulating tryptophan genes in Pseudomonadota, but not in Bacteroidota. However, the Bacteroid-trp RNA motif likely operates via the same mechanism of attenuation.

The Downstream-peptide motif refers to a conserved RNA structure identified by bioinformatics in the cyanobacterial genera Synechococcus and Prochlorococcus and one phage that infects such bacteria. It was also detected in marine samples of DNA from uncultivated bacteria, which are presumably other species of cyanobacteria.
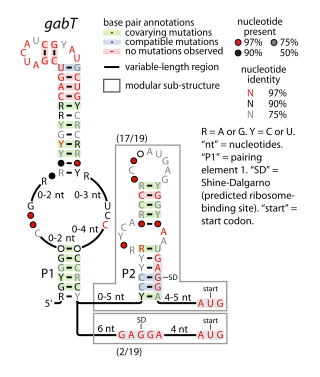
The gabT RNA motif is the name of a conserved RNA structure identified by bioinformatics whose function is unknown. The gabT motif has been detected exclusively in bacteria within the genus Pseudomonas, and is found only upstream of gabT genes, and downstream to gabD genes.

The gyrA RNA motif is a conserved RNA structure identified by bioinformatics. The RNAs are present in multiple species of bacteria within the order Pseudomonadales. This order contains the genus Pseudomonas, which includes the opportunistic human pathogen Pseudomonas aeruginosa and Pseudomonas syringae, a plant pathogen.

The L17 downstream element RNA motif is a conserved RNA structure identified in bacteria by bioinformatics. All known L17 downstream elements were detected immediately downstream of genes encoding the L17 subunit of the ribosome, and therefore might be in the 3' untranslated regions of these genes. The element is found in a variety of lactic acid bacteria and in the genus Listeria.

The Lacto-rpoB RNA motif is a conserved RNA structure identified by bioinformatics. It has been detected only in lactic acid bacteria, and is always located in the presumed 5' untranslated regions of rpoB genes. These genes encode a subunit of RNA polymerase, and it is hypothesized that Lacto-rpoB RNA participate in the regulation of these genes.
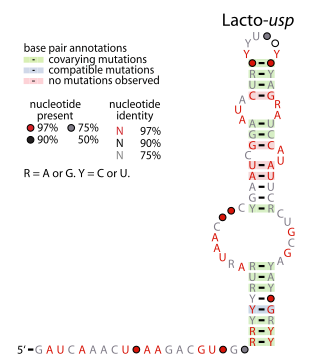
The Lacto-usp RNA motif is a conserved RNA structure identified in bacteria by bioinformatics. Lacto-usp RNAs are found exclusively in lactic acid bacteria, and exclusively in the possible 5′ untranslated regions of operons that contain a hypothetical gene and a usp gene. The usp gene encodes the universal stress protein. It was proposed that the Lacto-usp might correspond to the 6S RNA of the relevant species, because four of five of these species lack a predicted 6S RNA, and 6S RNAs commonly occur in 5′ UTRs of usp genes. However, given that the Lacto-usp RNA motif is much shorter than the standard 6S RNA structure, the function of Lacto-usp RNAs remains unclear.
The Lnt RNA motif refers to a conserved RNA structure found in certain bacteria. Specifically, Lnt RNAs are known only in species within the phylum Chlorobiota, and are located in the possible 5' untranslated regions of genes that are annotated as encoding apolipoprotein N-acyltransferase enzymes. There is some doubt as to whether the indicated motif is transcribed as RNA, or whether its reverse complement is transcribed. If the reverse complement is transcribed it would potentially in 5' UTRs of genes encoding bacteriochlorophyll A, and would be close to the start codon of those genes.
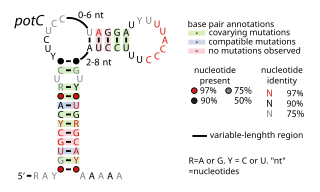
The potC RNA motif is a conserved RNA structure discovered using bioinformatics. The RNA is detected only in genome sequences derived from DNA that was extracted from uncultivated marine bacteria. Thus, this RNA is present in environmental samples, but not yet found in any cultivated organism. potC RNAs are located in the presumed 5' untranslated regions of genes predicted to encode either membrane transport proteins or peroxiredoxins. Therefore, it was hypothesized that potC RNAs are cis-regulatory elements, but their detailed function is unknown.

The Pseudomon-Rho RNA motif refers to a conserved RNA structure that was discovered using bioinformatics. The RNAs that conform to this motif are found in species within the genus Pseudomonas, as well as the related Azotobacter vinelandii. They are consistently located in what could be the 5' untranslated regions of genes that encode the Rho factor protein, and this arrangement in bacteria suggested that Pseudomon-Rho RNAs might be cis-regulatory elements that regulate concentrations of the Rho protein.
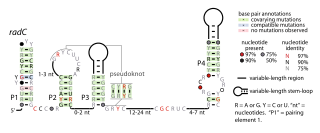
The radC RNA motif is a conserved RNA structure identified by bioinformatics. The radC RNA motif is found in certain bacteria where it is consistent located in the presumed 5' untranslated regions of genes whose encoded proteins bind DNA are interact with other proteins that bind DNA. These proteins include integrases, methyltransferases that might methylate DNA, proteins that inhibit restriction enzymes and radC genes. Although radC genes were thought to encode DNA repair proteins, this conclusion was based on mutation data that was later shown to affect a different gene. However, it is still possible that radC genes play some DNA-related role. No radC RNAs have been detected in any purified phage whose sequence was available as of 2010, although integrases are often used by phages.

The SAM-Chlorobi RNA motif is a conserved RNA structure that was identified by bioinformatics. The RNAs are found only in bacteria classified as within the phylum Chlorobiota. These RNAs are always in the 5' untranslated regions of operons that contain metK and ahcY genes. metK genes encode methionine adenosyltransferase, which synthesizes S-adenosyl methionine (SAM), and ahcY genes encode S-adenosylhomocysteine hydrolase, which degrade the related metabolite S-Adenosyl-L-homocysteine (SAH). In fact all predicted metK and ahcY genes within Chlorobiota bacteria as of 2010 are preceded by predicted SAM-Chlorobi RNAs. Predicted promoter sequences are consistently found upstream of SAM-Chlorobi RNAs, and these promoter sequences imply that SAM-Chlorobi RNAs are indeed transcribed as RNAs. The promoter sequences are commonly associated with strong transcription in the phyla Chlorobiota and Bacteroidota, but are not used by most lineages of bacteria. The placement of SAM-Chlorobi RNAs suggests that they are involved in the regulation of the metK/ahcY operon through an unknown mechanism.
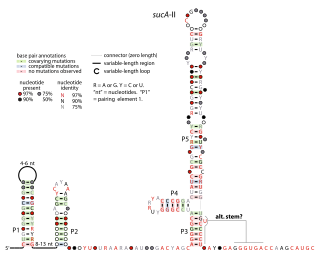
The sucA-II RNA motif is a conserved RNA structure identified by bioinformatics. It is consistently found in the presumed 5' untranslated regions of sucA genes, which encode Oxoglutarate dehydrogenase enzymes that participate in the citric acid cycle. Given this arrangement, sucA-II RNAs might regulate the downstream sucA gene. This genetic arrangement is similar to the previously reported sucA RNA motif. However, sucA-II RNAs are found only in bacteria classified within the genus Pseudomonas, whereas the previously reported motif is found only in betaproteobacteria.

The sucC RNA motif is a conserved RNA structure discovered using bioinformatics. sucC RNAs are found in the genus Pseudomonas. They ae consistently found in possible 5' untranslated regions of sucC genes. These genes encode Succinyl coenzyme A synthetase, and are hypothesised to be regulated by the sucC RNAs. sucC genes participate in the citric acid cycle, and another gene involved in the citric acid cycle, sucA, is also predicted to be regulated by a conserved RNA structure.

The Termite-flg RNA motif is a conserved RNA structure identified by bioinformatics. Genomic sequences corresponding to Termite-flg RNAs have been identified only in uncultivated bacteria present in the termite hindgut. As of 2010 it has not been identified in the DNA of any cultivated species, and is thus an example of RNAs present in environmental samples.

The yjdF RNA motif is a conserved RNA structure identified using bioinformatics. Most yjdF RNAs are located in bacteria classified within the phylum Bacillota. A yjdF RNA is found in the presumed 5' untranslated region of the yjdF gene in Bacillus subtilis, and almost all yjdF RNAs are found in the 5' UTRs of homologs of this gene. The function of the yjdF gene is unknown, but the protein that it is predicted to encode is classified by the Pfam Database as DUF2992.

Tetrahydrofolate riboswitches are a class of homologous RNAs in certain bacteria that bind tetrahydrofolate (THF). It is almost exclusively located in the probable 5' untranslated regions of protein-coding genes, and most of these genes are known to encode either folate transporters or enzymes involved in folate metabolism. For these reasons it was inferred that the RNAs function as riboswitches. THF riboswitches are found in a variety of Bacillota, specifically the orders Clostridiales and Lactobacillales, and more rarely in other lineages of bacteria. The THF riboswitch was one of many conserved RNA structures found in a project based on comparative genomics. The 3-d structure of the tetrahydrofolate riboswitch has been solved by separate groups using X-ray crystallography. These structures were deposited into the Protein Data Bank under accessions 3SD1 and 3SUX, with other entries containing variants.
The bacterial, archaeal and plant plastid code is the DNA code used by bacteria, archaea, prokaryotic viruses and chloroplast proteins. It is essentially the same as the standard code, however there are some variations in alternative start codons.


















