Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, research, and forensics. Commonly made in the laboratory by solid-phase chemical synthesis, these small bits of nucleic acids can be manufactured as single-stranded molecules with any user-specified sequence, and so are vital for artificial gene synthesis, polymerase chain reaction (PCR), DNA sequencing, molecular cloning and as molecular probes. In nature, oligonucleotides are usually found as small RNA molecules that function in the regulation of gene expression, or are degradation intermediates derived from the breakdown of larger nucleic acid molecules.

A DNA microarray is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or to genotype multiple regions of a genome. Each DNA spot contains picomoles of a specific DNA sequence, known as probes. These can be a short section of a gene or other DNA element that are used to hybridize a cDNA or cRNA sample under high-stringency conditions. Probe-target hybridization is usually detected and quantified by detection of fluorophore-, silver-, or chemiluminescence-labeled targets to determine relative abundance of nucleic acid sequences in the target. The original nucleic acid arrays were macro arrays approximately 9 cm × 12 cm and the first computerized image based analysis was published in 1981. It was invented by Patrick O. Brown. An example of its application is in SNPs arrays for polymorphisms in cardiovascular diseases, cancer, pathogens and GWAS analysis. It is also used for the identification of structural variations and the measurement of gene expression.

Fluorescein is an organic compound and dye based on the xanthene tricyclic structural motif, formally belonging to triarylmethine dyes family. It is available as a dark orange/red powder slightly soluble in water and alcohol. It is widely used as a fluorescent tracer for many applications.
A fluorophore is a fluorescent chemical compound that can re-emit light upon light excitation. Fluorophores typically contain several combined aromatic groups, or planar or cyclic molecules with several π bonds.
Fam or FAM is a colloquial term for 'family and friend' or an acronym of 'friend and mate' especially for intimate friends. It may also refer to:
In molecular biology, a hybridization probe(HP) is a fragment of DNA or RNA of usually 15–10000 nucleotide long which can be radioactively or fluorescently labeled. HP can be used to detect the presence of nucleotide sequences in analyzed RNA or DNA that are complementary to the sequence in the probe. The labeled probe is first denatured (by heating or under alkaline conditions such as exposure to sodium hydroxide) into single stranded DNA (ssDNA) and then hybridized to the target ssDNA (Southern blotting) or RNA (northern blotting) immobilized on a membrane or in situ.

Periodic acid–Schiff (PAS) is a staining method used to detect polysaccharides such as glycogen, and mucosubstances such as glycoproteins, glycolipids and mucins in tissues. The reaction of periodic acid oxidizes the vicinal diols in these sugars, usually breaking up the bond between two adjacent carbons not involved in the glycosidic linkage or ring closure in the ring of the monosaccharide units that are parts of the long polysaccharides, and creating a pair of aldehydes at the two free tips of each broken monosaccharide ring. The oxidation condition has to be sufficiently regulated so as to not oxidize the aldehydes further. These aldehydes then react with the Schiff reagent to give a purple-magenta color. A suitable basic stain is often used as a counterstain.

The southwestern blot, is a lab technique that involves identifying as well as characterizing DNA-binding proteins by their ability to bind to specific oligonucleotide probes. Determination of molecular weight of proteins binding to DNA is also made possible by the technique. The name originates from a combination of ideas underlying Southern blotting and Western blotting techniques of which they detect DNA and protein respectively. Similar to other types of blotting, proteins are separated by SDS-PAGE and are subsequently transferred to nitrocellulose membranes. Thereafter southwestern blotting begins to vary with regards to procedure as since the first blotting’s, many more have been proposed and discovered with goals of enhancing results. Former protocols were hampered by the need for large amounts of proteins and their susceptibility to degradation while being isolated.

An electrophoretic mobility shift assay (EMSA) or mobility shift electrophoresis, also referred as a gel shift assay, gel mobility shift assay, band shift assay, or gel retardation assay, is a common affinity electrophoresis technique used to study protein–DNA or protein–RNA interactions. This procedure can determine if a protein or mixture of proteins is capable of binding to a given DNA or RNA sequence, and can sometimes indicate if more than one protein molecule is involved in the binding complex. Gel shift assays are often performed in vitro concurrently with DNase footprinting, primer extension, and promoter-probe experiments when studying transcription initiation, DNA gang replication, DNA repair or RNA processing and maturation, as well as pre-mRNA splicing. Although precursors can be found in earlier literature, most current assays are based on methods described by Garner and Revzin and Fried and Crothers.
In molecular biology and genetics, the sense of a nucleic acid molecule, particularly of a strand of DNA or RNA, refers to the nature of the roles of the strand and its complement in specifying a sequence of amino acids. Depending on the context, sense may have slightly different meanings. For example, negative-sense strand of DNA is equivalent to the template strand, whereas the positive-sense strand is the non-template strand whose nucleotide sequence is equivalent to the sequence of the mRNA transcript.

6-Carboxyfluorescein (6-FAM) is a fluorescent dye with an absorption wavelength of 495 nm and an emission wavelength of 517 nm. A carboxyfluorescein molecule is a fluorescein molecule with a carboxyl group added. They are commonly used as a tracer agents. It is used in the sequencing of nucleic acids and in the labeling of nucleotides.
In biology, a branched DNA assay is a signal amplification assay that is used to detect nucleic acid molecules.
A dark quencher is a substance that absorbs excitation energy from a fluorophore and dissipates the energy as heat; while a typical (fluorescent) quencher re-emits much of this energy as light. Dark quenchers are used in molecular biology in conjunction with fluorophores. When the two are close together, such as in a molecule or protein, the fluorophore's emission is suppressed. This effect can be used to study molecular geometry and motion.
SNP genotyping is the measurement of genetic variations of single nucleotide polymorphisms (SNPs) between members of a species. It is a form of genotyping, which is the measurement of more general genetic variation. SNPs are one of the most common types of genetic variation. An SNP is a single base pair mutation at a specific locus, usually consisting of two alleles. SNPs are found to be involved in the etiology of many human diseases and are becoming of particular interest in pharmacogenetics. Because SNPs are conserved during evolution, they have been proposed as markers for use in quantitative trait loci (QTL) analysis and in association studies in place of microsatellites. The use of SNPs is being extended in the HapMap project, which aims to provide the minimal set of SNPs needed to genotype the human genome. SNPs can also provide a genetic fingerprint for use in identity testing. The increase of interest in SNPs has been reflected by the furious development of a diverse range of SNP genotyping methods.
Oligonucleotide synthesis is the chemical synthesis of relatively short fragments of nucleic acids with defined chemical structure (sequence). The technique is extremely useful in current laboratory practice because it provides a rapid and inexpensive access to custom-made oligonucleotides of the desired sequence. Whereas enzymes synthesize DNA and RNA only in a 5' to 3' direction, chemical oligonucleotide synthesis does not have this limitation, although it is most often carried out in the opposite, 3' to 5' direction. Currently, the process is implemented as solid-phase synthesis using phosphoramidite method and phosphoramidite building blocks derived from protected 2'-deoxynucleosides, ribonucleosides, or chemically modified nucleosides, e.g. LNA or BNA.
The DyLight Fluor family of fluorescent dyes are produced by Dyomics in collaboration with Thermo Fisher Scientific. DyLight dyes are typically used in biotechnology and research applications as biomolecule, cell and tissue labels for fluorescence microscopy, cell biology or molecular biology.
Κ-casein, or kappa casein, is a mammalian milk protein involved in several important physiological processes. Chymosin splits K-casein into an insoluble peptide and water-soluble glycomacropeptide (GMP). GMP is responsible for an increased efficiency of digestion, prevention of neonate hypersensitivity to ingested proteins, and inhibition of gastric pathogens. The human gene for κ-casein is CSN3.

MAGIChips, also known as "microarrays of gel-immobilized compounds on a chip" or "three-dimensional DNA microarrays", are devices for molecular hybridization produced by immobilizing oligonucleotides, DNA, enzymes, antibodies, and other compounds on a photopolymerized micromatrix of polyacrylamide gel pads of 100x100x20µm or smaller size. This technology is used for analysis of nucleic acid hybridization, specific binding of DNA, and low-molecular weight compounds with proteins, and protein-protein interactions.
Polony sequencing is an inexpensive but highly accurate multiplex sequencing technique that can be used to “read” millions of immobilized DNA sequences in parallel. This technique was first developed by Dr. George Church's group at Harvard Medical School. Unlike other sequencing techniques, Polony sequencing technology is an open platform with freely downloadable, open source software and protocols. Also, the hardware of this technique can be easily set up with a commonly available epifluorescence microscopy and a computer-controlled flowcell/fluidics system. Polony sequencing is generally performed on paired-end tags library that each molecule of DNA template is of 135 bp in length with two 17–18 bp paired genomic tags separated and flanked by common sequences. The current read length of this technique is 26 bases per amplicon and 13 bases per tag, leaving a gap of 4–5 bases in each tag.
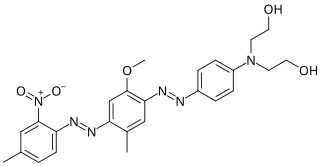
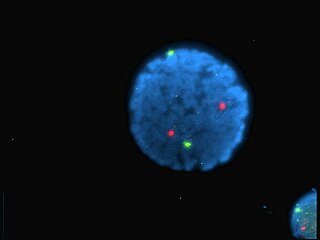
Black Hole Quencher 1 (BHQ1) is an example of dark quencher, which is used to quench green and yellow dyes, such as 6-carboxyfluorescein (6-FAM), tetrachlorofluorescein (TET), and hexachlorofluorescein (HEX). The role of quenchers is to absorb energy from a fluorophore and to re-emit the energy in the form of either heat or visible light. The absorption range of BHQ1 is from 480 to 580 nm with maximum absorption at 534 nm.









