Peptidoglycan or murein is a unique large macromolecule, a polysaccharide, consisting of sugars and amino acids that forms a mesh-like peptidoglycan layer outside the plasma membrane, the rigid cell wall characteristic of most bacteria. The sugar component consists of alternating residues of β-(1,4) linked N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM). Attached to the N-acetylmuramic acid is an oligopeptide chain made of three to five amino acids. The peptide chain can be cross-linked to the peptide chain of another strand forming the 3D mesh-like layer. Peptidoglycan serves a structural role in the bacterial cell wall, giving structural strength, as well as counteracting the osmotic pressure of the cytoplasm. This repetitive linking results in a dense peptidoglycan layer which is critical for maintaining cell form and withstanding high osmotic pressures, and it is regularly replaced by peptidoglycan production. Peptidoglycan hydrolysis and synthesis are two processes that must occur in order for cells to grow and multiply, a technique carried out in three stages: clipping of current material, insertion of new material, and re-crosslinking of existing material to new material.
Autolysins are endogenous lytic enzymes that break down the peptidoglycan components of biological cells which enables the separation of daughter cells following cell division. They are involved in cell growth, cell wall metabolism, cell division and separation, as well as peptidoglycan turnover and have similar functions to lysozymes.
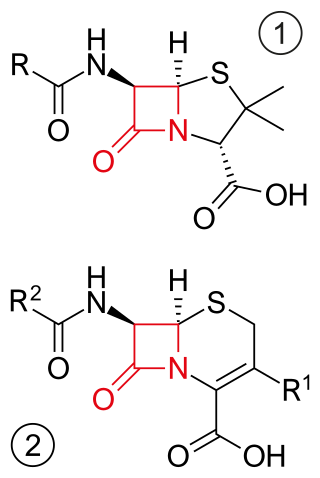
β-lactam antibiotics are antibiotics that contain a beta-lactam ring in their chemical structure. This includes penicillin derivatives (penams), cephalosporins and cephamycins (cephems), monobactams, carbapenems and carbacephems. Most β-lactam antibiotics work by inhibiting cell wall biosynthesis in the bacterial organism and are the most widely used group of antibiotics. Until 2003, when measured by sales, more than half of all commercially available antibiotics in use were β-lactam compounds. The first β-lactam antibiotic discovered, penicillin, was isolated from a strain of Penicillium rubens.
Braun's lipoprotein, found in some gram-negative cell walls, is one of the most abundant membrane proteins; its molecular weight is about 7.2 kDa. It is bound at its C-terminal end by a covalent bond to the peptidoglycan layer and is embedded in the outer membrane by its hydrophobic head. BLP tightly links the two layers and provides structural integrity to the outer membrane.

DD-transpeptidase is a bacterial enzyme that catalyzes the transfer of the R-L-αα-D-alanyl moiety of R-L-αα-D-alanyl-D-alanine carbonyl donors to the γ-OH of their active-site serine and from this to a final acceptor. It is involved in bacterial cell wall biosynthesis, namely, the transpeptidation that crosslinks the peptide side chains of peptidoglycan strands.

N-Acetylmuramic acid is an organic compound with the chemical formula C
11H
19NO
8. It is a monomer of peptidoglycan in most bacterial cell walls, which is built from alternating units of N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid, cross-linked by oligopeptides at the lactic acid residue of MurNAc.
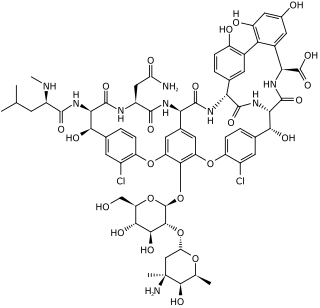
Glycopeptide antibiotics are a class of drugs of microbial origin that are composed of glycosylated cyclic or polycyclic nonribosomal peptides. Significant glycopeptide antibiotics include the anti-infective antibiotics vancomycin, teicoplanin, telavancin, ramoplanin and decaplanin, corbomycin, complestatin and the antitumor antibiotic bleomycin. Vancomycin is used if infection with methicillin-resistant Staphylococcus aureus (MRSA) is suspected.

Pseudopeptidoglycan is a major cell wall component of some Archaea that differs from bacterial peptidoglycan in chemical structure, but resembles bacterial peptidoglycan in function and physical structure. Pseudopeptidoglycan, in general, is only present in a few methanogenic archaea. The basic components are N-acetylglucosamine and N-acetyltalosaminuronic acid, which are linked by β-1,3-glycosidic bonds.
Penicillin-binding proteins (PBPs) are a group of proteins that are characterized by their affinity for and binding of penicillin. They are a normal constituent of many bacteria; the name just reflects the way by which the protein was discovered. All β-lactam antibiotics bind to PBPs, which are essential for bacterial cell wall synthesis. PBPs are members of a subgroup of enzymes called transpeptidases. Specifically, PBPs are DD-transpeptidases.

Lysins, also known as endolysins or murein hydrolases, are hydrolytic enzymes produced by bacteriophages in order to cleave the host's cell wall during the final stage of the lytic cycle. Lysins are highly evolved enzymes that are able to target one of the five bonds in peptidoglycan (murein), the main component of bacterial cell walls, which allows the release of progeny virions from the lysed cell. Cell-wall-containing Archaea are also lysed by specialized pseudomurein-cleaving lysins, while most archaeal viruses employ alternative mechanisms. Similarly, not all bacteriophages synthesize lysins: some small single-stranded DNA and RNA phages produce membrane proteins that activate the host's autolytic mechanisms such as autolysins.
In enzymology, glutamate racemase is an enzyme that catalyzes the chemical reaction
Peptidoglycan glycosyltransferase is an enzyme used in the biosynthesis of peptidoglycan. It transfers a disaccharide-peptide from a donor substrate to synthesize a glycan chain.
Zinc D-Ala-D-Ala carboxypeptidase (EC 3.4.17.14, Zn2+ G peptidase, D-alanyl-D-alanine hydrolase, D-alanyl-D-alanine-cleaving carboxypeptidase, DD-carboxypeptidase, G enzyme, DD-carboxypeptidase-transpeptidase) is an enzyme. This enzyme catalyses the following chemical reaction
Sortases are membrane anchored enzyme that sort these surface proteins onto the bacterial cell surface and anchor them to the peptidoglycan. There are different types of sortases and each catalyse the anchoring of different proteins to cell walls.

Lipid II is a precursor molecule in the synthesis of the cell wall of bacteria. It is a peptidoglycan, which is amphipathic and named for its bactoprenol hydrocarbon chain, which acts as a lipid anchor, embedding itself in the bacterial cell membrane. Lipid II must translocate across the cell membrane to deliver and incorporate its disaccharide-pentapeptide "building block" into the peptidoglycan mesh. Lipid II is the target of several antibiotics.

The divisome is a protein complex in bacteria that is responsible for cell division, constriction of inner and outer membranes during division, and peptidoglycan (PG) synthesis at the division site. The divisome is a membrane protein complex with proteins on both sides of the cytoplasmic membrane. In gram-negative cells it is located in the inner membrane. The divisome is nearly ubiquitous in bacteria although its composition may vary between species.
Renee Elizabeth Sockett is a professor and microbiologist in the School of Life Sciences at the University of Nottingham. She is a world-leading expert on Bdellovibrio bacteriovorus, a species of predatory bacteria.
D-Amino acids are amino acids where the stereogenic carbon alpha to the amino group has the D-configuration. For most naturally-occurring amino acids, this carbon has the L-configuration. D-Amino acids are occasionally found in nature as residues in proteins. They are formed from ribosomally-derived D-amino acid residues.
Corbomycin is a member of the glycopeptide family of antibiotics that are produced by soil bacteria.

Joshua Shaevitz is an American biophysicist and Professor of Physics at the Lewis-Sigler Institute at Princeton University in Princeton, NJ. He is known for his work in single-molecule biophysics, bacterial growth and motility, and animal behavior.












