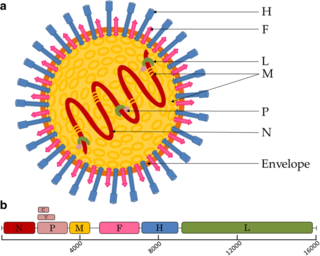
Paramyxoviridae is a family of negative-strand RNA viruses in the order Mononegavirales. Vertebrates serve as natural hosts. Diseases associated with this family include measles, mumps, and respiratory tract infections. The family has four subfamilies, 17 genera, and 78 species, three genera of which are unassigned to a subfamily.

Orthohantavirus is a genus of single-stranded, enveloped, negative-sense RNA viruses in the family Hantaviridae within the order Bunyavirales. Members of this genus may be called orthohantaviruses or simply hantaviruses.

Influenza A virus (IAV) is a pathogen that causes the flu in birds and some mammals, including humans. It is an RNA virus whose subtypes have been isolated from wild birds. Occasionally, it is transmitted from wild to domestic birds, and this may cause severe disease, outbreaks, or human influenza pandemics.

Antigenic shift is the process by which two or more different strains of a virus, or strains of two or more different viruses, combine to form a new subtype having a mixture of the surface antigens of the two or more original strains. The term is often applied specifically to influenza, as that is the best-known example, but the process is also known to occur with other viruses, such as visna virus in sheep. Antigenic shift is a specific case of reassortment or viral shift that confers a phenotypic change.

Influenza A virus subtype H5N1 (A/H5N1) is a subtype of the influenza A virus which can cause illness in humans and many other species. A bird-adapted strain of H5N1, called HPAI A(H5N1) for highly pathogenic avian influenza virus of type A of subtype H5N1, is the highly pathogenic causative agent of H5N1 flu, commonly known as avian influenza. It is enzootic in many bird populations, especially in Southeast Asia. One strain of HPAI A(H5N1) is spreading globally after first appearing in Asia. It is epizootic and panzootic, killing tens of millions of birds and spurring the culling of hundreds of millions of others to stem its spread. Many references to "bird flu" and H5N1 in the popular media refer to this strain.

Bunyavirales is an order of segmented negative-strand RNA viruses with mainly tripartite genomes. Member viruses infect arthropods, plants, protozoans, and vertebrates. It is the only order in the class Ellioviricetes. The name Bunyavirales derives from Bunyamwera, where the original type species Bunyamwera orthobunyavirus was first discovered. Ellioviricetes is named in honor of late virologist Richard M. Elliott for his early work on bunyaviruses.

Swine influenza is an infection caused by any of several types of swine influenza viruses. Swine influenza virus (SIV) or swine-origin influenza virus (S-OIV) refers to any strain of the influenza family of viruses that is endemic in pigs. As of 2009, identified SIV strains include influenza C and the subtypes of influenza A known as H1N1, H1N2, H2N1, H3N1, H3N2, and H2N3.

Oropouche fever is a tropical viral infection transmitted by biting midges and mosquitoes from the blood of sloths to humans. This disease is named after the region where it was first discovered and isolated at the Trinidad Regional Virus Laboratory in 1955 by the Oropouche River in Trinidad and Tobago. Oropouche fever is caused by a specific arbovirus, the Oropouche virus (OROV), of the Bunyaviridae family.

Influenza A virus subtype H3N2 (A/H3N2) is a subtype of viruses that causes influenza (flu). H3N2 viruses can infect birds and mammals. In birds, humans, and pigs, the virus has mutated into many strains. In years in which H3N2 is the predominant strain, there are more hospitalizations.

H5N1 genetic structure is the molecular structure of the H5N1 virus's RNA.

The Argentine brown bat, is a bat species from South and Central America.

Andes orthohantavirus (ANDV), a species of Orthohantavirus, is a major causative agent of hantavirus cardiopulmonary syndrome (HCPS) and hantavirus pulmonary syndrome (HPS) in South America. It is named for the Andes mountains of Chile and Argentina, where it was first discovered. Originating in the reservoir of rodents, Andes orthohantavirus is easily transmitted to humans who come into contact with infected rodents or their fecal droppings. However, infected rodents do not appear ill, so there is no readily apparent indicator to determine whether the rodent is infected or not. Additionally, Andes orthohantavirus, specifically, is the only hantavirus that can be spread by human to human contact via bodily fluids or long-term contact from one infected individual to a healthy person.

Fujian flu refers to flu caused by either a Fujian human flu strain of the H3N2 subtype of the Influenza A virus or a Fujian bird flu strain of the H5N1 subtype of the Influenza A virus. These strains are named after Fujian, a coastal province in Southeast China.

Influenza, commonly known as "the flu", is an infectious disease caused by influenza viruses. Symptoms range from mild to severe and often include fever, runny nose, sore throat, muscle pain, headache, coughing, and fatigue. These symptoms begin from one to four days after exposure to the virus and last for about 2–8 days. Diarrhea and vomiting can occur, particularly in children. Influenza may progress to pneumonia, which can be caused by the virus or by a subsequent bacterial infection. Other complications of infection include acute respiratory distress syndrome, meningitis, encephalitis, and worsening of pre-existing health problems such as asthma and cardiovascular disease.

The pandemic H1N1/09 virus is a swine origin influenza A virus subtype H1N1 strain that was responsible for the 2009 swine flu pandemic. This strain is often called swine flu by the public media. For other names, see the Nomenclature section below.
Batai orthobunyavirus (BATV) is a RNA virus belonging to order Bunyavirales, genus Orthobunyavirus.
Cache Valley orthobunyavirus (CVV) is a member of the order Bunyavirales, genus Orthobunyavirus, and serogroup Bunyamwera, which was first isolated in 1956 from Culiseta inornata mosquitos collected in Utah's Cache Valley. CVV is an enveloped arbovirus, nominally 80–120 nm in diameter, whose genome is composed of three single-stranded, negative-sense RNA segments. The large segment of related bunyaviruses is approximately 6800 bases in length and encodes a probable viral polymerase. The middle CVV segment has a 4463-nucleotide sequence and the smallest segment encodes for the nucleocapsid, and a second non-structural protein. CVV has been known to cause outbreaks of spontaneous abortion and congenital malformations in ruminants such as sheep and cattle. CVV rarely infects humans, but when they are infected it has caused encephalitis and multiorgan failure.
This glossary of virology is a list of definitions of terms and concepts used in virology, the study of viruses, particularly in the description of viruses and their actions. Related fields include microbiology, molecular biology, and genetics.

Orthornavirae is a kingdom of viruses that have genomes made of ribonucleic acid (RNA), including genes which encode an RNA-dependent RNA polymerase (RdRp). The RdRp is used to transcribe the viral RNA genome into messenger RNA (mRNA) and to replicate the genome. Viruses in this kingdom share a number of characteristics which promote rapid evolution, including high rates of genetic mutation, recombination, and reassortment.
Ngari virus (NRIV) is a single-stranded, negative sense, tri-segmented RNA virus. It is a subtype of the Bunyamwera virus (BUNV) and closely related to the Batai virus (BATV). NRIV is the only reassortment virus of the subtypes. There is evidence suggesting that NRIV stems from a naturally occurring reassortment event in which a host was infected with both BUNV and BATV. It is commonly found in areas that experience an outbreak of Rift Valley fever virus (RFVF)













