HNH Endonuclease-Associated RNA and ORF (HEARO) RNAs conform to a conserved RNA structure that was identified in bacteria by bioinformatics. HEARO RNAs average roughly 300 nucleotides, which is comparable to the size of many ribozymes, which catalyze chemical reactions.

A wide variety of non-coding RNAs have been identified in various species of organisms known to science. However, RNAs have also been identified in "metagenomics" sequences derived from samples of DNA or RNA extracted from the environment, which contain unknown species. Initial work in this area detected homologs of known bacterial RNAs in such metagenome samples. Many of these RNA sequences were distinct from sequences within cultivated bacteria, and provide the potential for additional information on the RNA classes to which they belong.
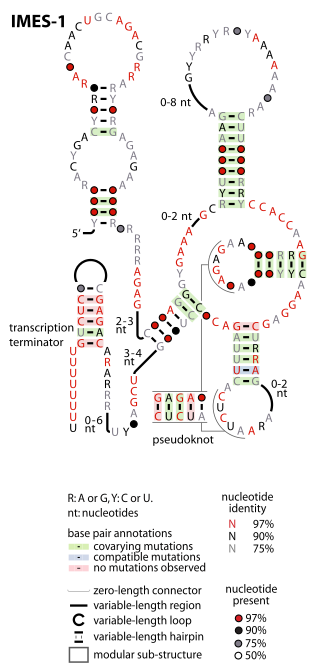
The IMES-1 RNA motif is a conserved RNA structure that was identified in marine environmental sequences by two studies based on metagenomics and bioinformatics, the first analyzing metatranscriptome (RNA) data and the second using metagenome (DNA) data. These RNAs are present in environmental sequences, and as of 2009 are not known to be present in any cultivated species. However, the species that use these RNAs are most closely related to known alphaproteobacteria and gammaproteobacteria. IMES-1 RNAs make up a significant portion of marine RNA transcripts and are exceptionally abundant in that over five times as many IMES-1 RNAs were found as ribosomes in RNAs sampled from the Pacific Ocean. Only two bacterial RNAs are known to be more highly transcribed than ribosomes. IMES-1 RNAs were also detected in abundance in Block Island Sound in the Atlantic Ocean.
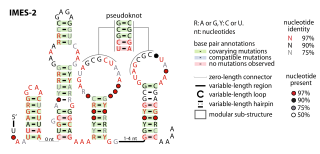
The IMES-2 RNA motif is a conserved RNA structure that was identified by a study based on metagenomics and bioinformatics, and the underlying RNA sequences were identified independently by a similar earlier study. These RNAs are present in environmental sequences, and when discovered were not known to be present in any cultivated species. However, an IMES-2 RNA has been detected in alphaproteobacterium HIMB114, which is classified in the SAR11 clade of marine bacteria. This finding fits with earlier predictions that species that use IMES-2 RNAs are most closely related to alphaproteobacteria. IMES-2 RNAs are exceptionally abundant, as twice as many IMES-2 RNAs were found as ribosomes in RNAs sampled from the Pacific Ocean. Only two bacterial RNAs are known to be more highly transcribed than ribosomes.

The IMES-3 RNA motif is a conserved RNA structure that was identified based on metagenomics and bioinformatics, and the underlying RNA sequences were identified independently by an earlier study. These RNAs are present in environmental sequences, and as of 2009 are not known to be present in any cultivated species. IMES-3 RNAs are abundant in comparison to ribosomes in RNAs sampled from the Pacific Ocean.
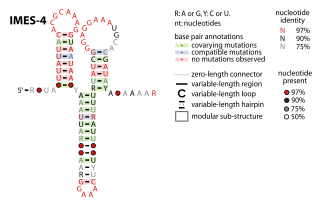
The IMES-4 RNA motif is a conserved RNA structure that was identified in marine environmental sequences by metagenomics and bioinformatics. These RNAs are present in environmental sequences, and as of 2009 are not known to be present in any cultivated species. IMES-4 RNAs are fairly abundant in comparison to ribosomes in RNAs sampled from the Pacific Ocean.
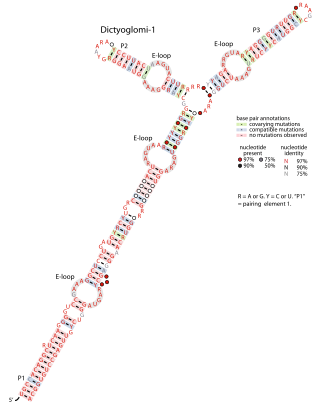
The Dictyoglomi-1 RNA motif is a conserved RNA structure that was discovered via bioinformatics. Only four instances of the RNA were detected, and all are in the bacterial phylum Dictyoglomota, whose members have not been extensively studied. The RNA might have a cis-regulatory role, but the evidence is ambiguous. Because of the few instances of Dictyoglomi-1 RNAs known, it is also unknown whether the RNA structure might extend further in the 5′ or 3′ direction, or in both directions.

The Gut-1 RNA motif is a conserved RNA structure identified by bioinformatics. These RNAs are present in environmental sequences, and as of 2010 are not known to be present in any species that has been grown under laboratory conditions. Gut-1 RNA is exclusively found in DNA from uncultivated bacteria present in samples from the human gut.

The manA RNA motif refers to a conserved RNA structure that was identified by bioinformatics. Instances of the manA RNA motif were detected in bacteria in the genus Photobacterium and phages that infect certain kinds of cyanobacteria. However, most predicted manA RNA sequences are derived from DNA collected from uncultivated marine bacteria. Almost all manA RNAs are positioned such that they might be in the 5' untranslated regions of protein-coding genes, and therefore it was hypothesized that manA RNAs function as cis-regulatory elements. Given the relative complexity of their secondary structure, and their hypothesized cis-regulatory role, they might be riboswitches.
The wcaG RNA motif is an RNA structure conserved in some bacteria that was detected by bioinformatics. wcaG RNAs are found in certain phages that infect cyanobacteria. Most known wcaG RNAs were found in sequences of DNA extracted from uncultivated marine bacteria. wcaG RNAs might function as cis-regulatory elements, in view of their consistent location in the possible 5' untranslated regions of genes. It was suggested the wcaG RNAs might further function as riboswitches.

PhotoRC RNA motifs refer to conserved RNA structures that are associated with genes acting in the photosynthetic reaction centre of photosynthetic bacteria. Two such RNA classes were identified and called the PhotoRC-I and PhotoRC-II motifs. PhotoRC-I RNAs were detected in the genomes of some cyanobacteria. Although no PhotoRC-II RNA has been detected in cyanobacteria, one is found in the genome of a purified phage that infects cyanobacteria. Both PhotoRC-I and PhotoRC-II RNAs are present in sequences derived from DNA that was extracted from uncultivated marine bacteria.
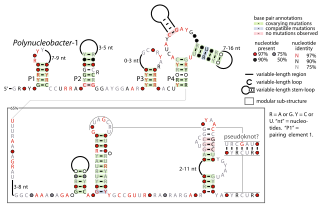
The Polynucleobacter-1 RNA motif is a conserved RNA structure that was identified by bioinformatics. The RNA structure is predominantly located in genome sequences derived from DNA extracted from uncultivated marine samples. However it was also predicted in the genome of Polynucleobacter species QLW-P1DMWA-1, a kind of betaproteobacteria. The RNAs are often located near to a conserved gene that might be homologous to a gene found in a phage that infects cyanobacteria. However, it is unknown if the RNA is used by phages.
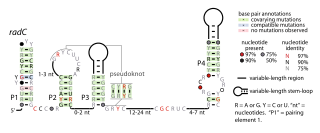
The radC RNA motif is a conserved RNA structure identified by bioinformatics. The radC RNA motif is found in certain bacteria where it is consistent located in the presumed 5' untranslated regions of genes whose encoded proteins bind DNA are interact with other proteins that bind DNA. These proteins include integrases, methyltransferases that might methylate DNA, proteins that inhibit restriction enzymes and radC genes. Although radC genes were thought to encode DNA repair proteins, this conclusion was based on mutation data that was later shown to affect a different gene. However, it is still possible that radC genes play some DNA-related role. No radC RNAs have been detected in any purified phage whose sequence was available as of 2010, although integrases are often used by phages.

The Termite-flg RNA motif is a conserved RNA structure identified by bioinformatics. Genomic sequences corresponding to Termite-flg RNAs have been identified only in uncultivated bacteria present in the termite hindgut. As of 2010 it has not been identified in the DNA of any cultivated species, and is thus an example of RNAs present in environmental samples.

The Whalefall-1 RNA motif refers to a conserved RNA structure that was discovered using bioinformatics. Structurally, the motif consists of two stem-loops, the second of which is often terminated by a CUUG tetraloop, which is an energetically favorable RNA sequence. Whalefall-1 RNAs are found only in DNA extracted from uncultivated bacteria found on whale fall, i.e., a whale carcass. As of 2010, Whalefall-1 RNAs have not been detected in any known, cultivated species of bacteria, and are thus one of several RNAs present in environmental samples.

RNAs Associated with Genes Associated with Twister and Hammerhead ribozymes (RAGATH) refers to a bioinformatics strategy that was devised to find self-cleaving ribozymes in bacteria. It also refers to candidate RNAs, or RAGATH RNA motifs, discovered using this strategy.
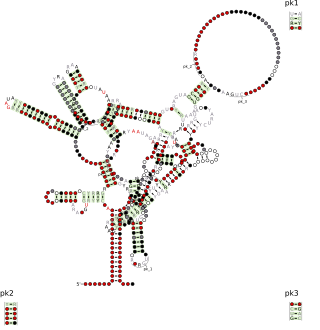
The Rumen-Originating, Ornate, Large (ROOL) RNA motif was originally discovered by bioinformatics by analyzing metagenomic sequences from cow rumen. ROOL RNAs are found in a variety of bacterial species and apparently do not code for proteins. The RNA has a complex RNA secondary structure and its average size of 581 nucleotides is unusually large for bacterial non-coding RNAs. This large size and structural complexity for a bacterial RNA is consistent with properties of large ribozymes.

The DUF2800 RNA motif is a conserved RNA structure that was discovered by bioinformatics. DUF2800 motif RNAs are found in Bacillota. DUF2800 RNAs are also predicted in the phyla Actinomycetota and Synergistota, although these RNAs are likely the result of recent horizontal gene transfer or conceivably sequence contamination.
The freshwater-2 RNA motif is a conserved RNA structure that was discovered by bioinformatics. Freshwater-2 motif RNAs are found in metagenomic sequences that are isolated from aquatic and especially freshwater environments. As of 2018, no freshwater-2 RNA has been identified in a classified organism.

The Ocean-VII RNA motif is a conserved RNA structure that was discovered by bioinformatics. Ocean-VII motifs are found in metagenomic sequences isolated from various marine environments, and are not yet known in any classified organism. This environmental context is similar to other marine RNAs that were found previously by predominantly bioinformatic or experimental methods.

















