The genotype of an organism is its complete set of genetic material. Genotype can also be used to refer to the alleles or variants an individual carries in a particular gene or genetic location. The number of alleles an individual can have in a specific gene depends on the number of copies of each chromosome found in that species, also referred to as ploidy. In diploid species like humans, two full sets of chromosomes are present, meaning each individual has two alleles for any given gene. If both alleles are the same, the genotype is referred to as homozygous. If the alleles are different, the genotype is referred to as heterozygous.

Heredity, also called inheritance or biological inheritance, is the passing on of traits from parents to their offspring; either through asexual reproduction or sexual reproduction, the offspring cells or organisms acquire the genetic information of their parents. Through heredity, variations between individuals can accumulate and cause species to evolve by natural selection. The study of heredity in biology is genetics.

In genetics, the phenotype is the set of observable characteristics or traits of an organism. The term covers the organism's morphology, its developmental processes, its biochemical and physiological properties, its behavior, and the products of behavior. An organism's phenotype results from two basic factors: the expression of an organism's genetic code and the influence of environmental factors. Both factors may interact, further affecting the phenotype. When two or more clearly different phenotypes exist in the same population of a species, the species is called polymorphic. A well-documented example of polymorphism is Labrador Retriever coloring; while the coat color depends on many genes, it is clearly seen in the environment as yellow, black, and brown. Richard Dawkins in 1978 and then again in his 1982 book The Extended Phenotype suggested that one can regard bird nests and other built structures such as caddisfly larva cases and beaver dams as "extended phenotypes".
Allele frequency, or gene frequency, is the relative frequency of an allele at a particular locus in a population, expressed as a fraction or percentage. Specifically, it is the fraction of all chromosomes in the population that carry that allele over the total population or sample size. Microevolution is the change in allele frequencies that occurs over time within a population.

In biology, polymorphism is the occurrence of two or more clearly different morphs or forms, also referred to as alternative phenotypes, in the population of a species. To be classified as such, morphs must occupy the same habitat at the same time and belong to a panmictic population.

A haplotype is a group of alleles in an organism that are inherited together from a single parent.

Wied's marmoset, also known as Wied's black-tufted-ear marmoset, is a New World monkey that lives in tropical and subtropical forests of eastern Brazil. Unlike other marmosets, Wied's marmoset lives in groups consisting of 4 or 5 females and 2 or 3 males. They are matriarchal, and only the dominant female is allowed to mate. Like other marmosets, the offspring are always born in pairs.
Genetic load is the difference between the fitness of an average genotype in a population and the fitness of some reference genotype, which may be either the best present in a population, or may be the theoretically optimal genotype. The average individual taken from a population with a low genetic load will generally, when grown in the same conditions, have more surviving offspring than the average individual from a population with a high genetic load. Genetic load can also be seen as reduced fitness at the population level compared to what the population would have if all individuals had the reference high-fitness genotype. High genetic load may put a population in danger of extinction.

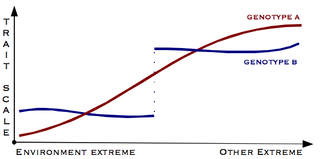
Gene–environment interaction is when two different genotypes respond to environmental variation in different ways. A norm of reaction is a graph that shows the relationship between genes and environmental factors when phenotypic differences are continuous. They can help illustrate GxE interactions. When the norm of reaction is not parallel, as shown in the figure below, there is a gene by environment interaction. This indicates that each genotype responds to environmental variation in a different way. Environmental variation can be physical, chemical, biological, behavior patterns or life events.
In ecology and genetics, a reaction norm, also called a norm of reaction, describes the pattern of phenotypic expression of a single genotype across a range of environments. One use of reaction norms is in describing how different species—especially related species—respond to varying environments. But differing genotypes within a single species may also show differing reaction norms relative to a particular phenotypic trait and environment variable. For every genotype, phenotypic trait, and environmental variable, a different reaction norm can exist; in other words, an enormous complexity can exist in the interrelationships between genetic and environmental factors in determining traits. The concept was introduced by Richard Woltereck in 1909.
Canalisation is a measure of the ability of a population to produce the same phenotype regardless of variability of its environment or genotype. It is a form of evolutionary robustness. The term was coined in 1942 by C. H. Waddington to capture the fact that "developmental reactions, as they occur in organisms submitted to natural selection...are adjusted so as to bring about one definite end-result regardless of minor variations in conditions during the course of the reaction". He used this word rather than robustness to consider that biological systems are not robust in quite the same way as, for example, engineered systems.
Genetic equilibrium is the condition of an allele or genotype in a gene pool where the frequency does not change from generation to generation. Genetic equilibrium describes a theoretical state that is the basis for determining whether and in what ways populations may deviate from it. Hardy–Weinberg equilibrium is one theoretical framework for studying genetic equilibrium. It is commonly studied using models that take as their assumptions those of Hardy-Weinberg, meaning:
In biology, a cline is a measurable gradient in a single characteristic of a species across its geographical range. Clines usually have a genetic, or phenotypic character. They can show either smooth, continuous gradation in a character, or more abrupt changes in the trait from one geographic region to the next.

Symbiodinium is a genus of dinoflagellates that encompasses the largest and most prevalent group of endosymbiotic dinoflagellates known and have photosymbiotic relationships with many species. These unicellular microalgae commonly reside in the endoderm of tropical cnidarians such as corals, sea anemones, and jellyfish, where the products of their photosynthetic processing are exchanged in the host for inorganic molecules. They are also harbored by various species of demosponges, flatworms, mollusks such as the giant clams, foraminifera (soritids), and some ciliates. Generally, these dinoflagellates enter the host cell through phagocytosis, persist as intracellular symbionts, reproduce, and disperse to the environment. The exception is in most mollusks, where these symbionts are intercellular. Cnidarians that are associated with Symbiodinium occur mostly in warm oligotrophic (nutrient-poor), marine environments where they are often the dominant constituents of benthic communities. These dinoflagellates are therefore among the most abundant eukaryotic microbes found in coral reef ecosystems.
Population genomics is the large-scale comparison of DNA sequences of populations. Population genomics is a neologism that is associated with population genetics. Population genomics studies genome-wide effects to improve our understanding of microevolution so that we may learn the phylogenetic history and demography of a population.

Anthothoe albocincta, or white-striped anemone, is a species of sea anemone in the family Sagartiidae. It is native to the coasts of Australia and New Zealand.

Community genetics is a recently emerged field in biology that fuses elements of community ecology, evolutionary biology, and molecular and quantitative genetics. Antonovics first articulated the vision for such a field, and Whitham et al. formalized its definition as "The study of the genetic interactions that occur between species and their abiotic environment in complex communities." The field aims to bridge the gaps in the study of evolution and ecology, within the multivariate community context in which ecological and evolutionary features are embedded. The documentary movie A Thousand Invisible Cords provides an introduction to the field and its implications.
Evolutionary rescue is a process by which a population—that would have gone extinct in the absence of evolution—persists due to natural selection acting on heritable variation. Coined by Gomulkiewicz & Holt in 1995, evolutionary rescue was described as a continuously changing environment predicted to appear as a stable lag of the mean trait value behind a moving environmental optimum, where the rate of evolution and change in the environment are equal. Evolutionary rescue is often confused with two other commonplace forms of rescue: genetic rescue and demographic rescue-in nature due to overlapping similarities. Figure 1 highlights the different pathways that result in their respective rescue.
This glossary of genetics and evolutionary biology is a list of definitions of terms and concepts used in the study of genetics and evolutionary biology, as well as sub-disciplines and related fields, with an emphasis on classical genetics, quantitative genetics, population biology, phylogenetics, speciation, and systematics. Overlapping and related terms can be found in Glossary of cellular and molecular biology, Glossary of ecology, and Glossary of biology.

Landscape genetics is the scientific discipline that combines population genetics and landscape ecology. It broadly encompasses any study that analyses plant or animal population genetic data in conjunction with data on the landscape features and matrix quality where the sampled population lives. This allows for the analysis of microevolutionary processes affecting the species in light of landscape spatial patterns, providing a more realistic view of how populations interact with their environments. Landscape genetics attempts to determine which landscape features are barriers to dispersal and gene flow, how human-induced landscape changes affect the evolution of populations, the source-sink dynamics of a given population, and how diseases or invasive species spread across landscapes.










