Biosynthesis, i.e., chemical synthesis occurring in biological contexts, is a term most often referring to multi-step, enzyme-catalyzed processes where chemical substances absorbed as nutrients serve as enzyme substrates, with conversion by the living organism either into simpler or more complex products. Examples of biosynthetic pathways include those for the production of amino acids, lipid membrane components, and nucleotides, but also for the production of all classes of biological macromolecules, and of acetyl-coenzyme A, adenosine triphosphate, nicotinamide adenine dinucleotide and other key intermediate and transactional molecules needed for metabolism. Thus, in biosynthesis, any of an array of compounds, from simple to complex, are converted into other compounds, and so it includes both the catabolism and anabolism of complex molecules. Biosynthetic processes are often represented via charts of metabolic pathways. A particular biosynthetic pathway may be located within a single cellular organelle, while others involve enzymes that are located across an array of cellular organelles and structures.

Clavulanic acid is a β-lactam drug that functions as a mechanism-based β-lactamase inhibitor. While not effective by itself as an antibiotic, when combined with penicillin-group antibiotics, it can overcome antibiotic resistance in bacteria that secrete β-lactamase, which otherwise inactivates most penicillins.
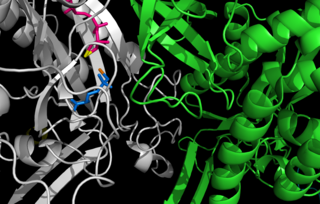
Glutamine synthetase (GS) is an enzyme that plays an essential role in the metabolism of nitrogen by catalyzing the condensation of glutamate and ammonia to form glutamine:
Chromosome 21 is one of the 23 pairs of chromosomes in humans. Chromosome 21 is both the smallest human autosome and chromosome, with 46.7 million base pairs representing about 1.5 percent of the total DNA in cells. Most people have two copies of chromosome 21, while those with three copies of chromosome 21 have Down syndrome.

Amino acid biosynthesis is the set of biochemical processes by which the amino acids are produced. The substrates for these processes are various compounds in the organism's diet or growth media. Not all organisms are able to synthesize all amino acids. For example, humans can synthesize 11 of the 20 standard amino acids. These 11 are called the non-essential amino acids.
Purine metabolism refers to the metabolic pathways to synthesize and break down purines that are present in many organisms.

Guanosine monophosphate synthetase, also known as GMPS is an enzyme that converts xanthosine monophosphate to guanosine monophosphate.
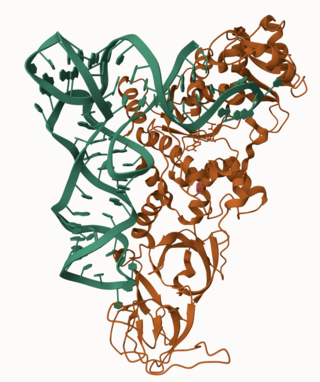
Glutamine—tRNA ligase or glutaminyl-tRNA synthetase (GlnRS) is an aminoacyl-tRNA synthetase, also called tRNA-ligase. is an enzyme that attaches the amino acid glutamine onto its cognate tRNA.
Glu-tRNAGln amidotransferase or glutaminyl-tRNA synthase (glutamine-hydrolysing) enzyme is an amidotransferase that catalyzes the conversion of the non-cognate amino acid glutamyl-tRNAGln to the cognate glutaminyl-tRNAGln.. It catalyzes the reaction:

Folylpolyglutamate synthase, mitochondrial is an enzyme that in humans is encoded by the FPGS gene.

The Downstream-peptide motif refers to a conserved RNA structure identified by bioinformatics in the cyanobacterial genera Synechococcus and Prochlorococcus and one phage that infects such bacteria. It was also detected in marine samples of DNA from uncultivated bacteria, which are presumably other species of cyanobacteria.

The glutamine riboswitch is a conserved RNA structure that was predicted by bioinformatics. It is present in a variety of lineages of cyanobacteria, as well as some phages that infect cyanobacteria. It is also found in DNA extracted from uncultivated bacteria living in the ocean that are presumably species of cyanobacteria.
Archaeosine synthase is an enzyme with systematic name L-glutamine:7-cyano-7-carbaguanine aminotransferase. This enzyme catalyses the following chemical reaction

The folP RNA motif is a conserved RNA structure that was discovered by bioinformatics. folP motifs are found in the genus Dialister.

The ilvH RNA motif is a conserved RNA structure that was discovered by bioinformatics. ilvH motifs are found in Betaproteobacteria.
The leuA-Halobacteria RNA motif is a conserved RNA structure that was discovered by bioinformatics. leuA-Halobacteria motifs are found in Halobacteriaceae, a lineage of archaea.
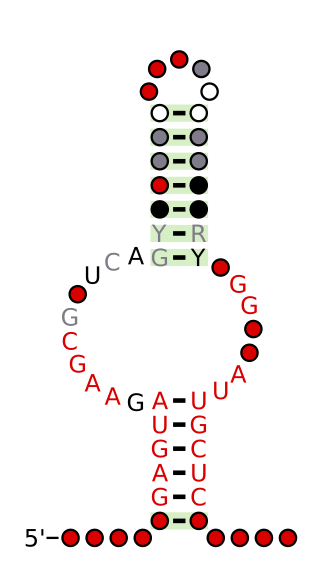
The NMT1 RNA motif is a conserved RNA structure that was discovered by bioinformatics. NMT1 motif RNAs are found in Pseudomonadota. There is also one NMT1 RNA in each of Bacteroidota and Actinomycetota, but these appear to be the result of recent horizontal gene transfer or sequence contamination before or during genome sequencing
A Streptomyces-metKH RNA motif is a conserved RNA structure that was discovered by bioinformatics. Such motifs are found in the genus Streptomyces, and are present upstream of either metK genes, which encode the S-adenosylmethionine synthetase enzyme or metH genes, which encodes the adenosylcobalamin-dependent form of methionine synthase. The RNA structures upstream of metK and metH genes are distinct from each other, but exhibit overall similar sequence and secondary structure features, suggesting that they are related to one another. Their presence upstream of protein-coding genes, and the fact that the genes perform related steps in metabolism, suggests that the RNAs function as cis-regulatory elements.

The sul1 RNA motif is a conserved RNA structure that was discovered by bioinformatics. Energetically stable tetraloops often occur in this motif. sul1 motif RNAs are found in Alphaproteobacteria.












